Articles
- Page Path
- HOME > J. Microbiol > Volume 63(1); 2025 > Article
-
Full article
Lactic acid bacteria from Ethiopian traditional beverage, Tella: technological and metabolic profiles for industrial application - Gashaw Assefa Yehuala1,2,3, Jaein Choe1, Nurelegne Tefera Shibeshi3, Kumsa Delessa3, Asnake Desalegn4, Mi-Kyung Park1
-
Journal of Microbiology 2025;63(1):e.2409008.
DOI: https://doi.org/10.71150/jm.2409008
Published online: January 24, 2025
1School of Food Science and Biotechnology, and Food and Bio-Industry Research Institute, Kyungpook National University, Daegu 41566, Republic of Korea
2Biotechnology and Bioprocess Center of Excellence, Addis Ababa Science and Technology University, Addis Ababa P.O. Box 16417, Ethiopia
3School of Chemical and Bio-Engineering, Addis Ababa Institute of Technology, Addis Ababa University, Addis Ababa P.O. Box 385, Ethiopia
4Department of Microbial, Cellular and Molecular Biology, College of Natural and Computational Science, Addis Ababa University, Addis Ababa P.O. Box 1176, Ethiopia
- Mi-Kyung Park parkmik@knu.ac.kr
© The Microbiological Society of Korea
This is an Open Access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/4.0) which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
ABSTRACT
- Tella is a traditional beverage widely accepted by consumers, despite the lack of product consistency owing to its reliance on natural fermentation. This study aimed to identify potential industrial lactic acid bacteria (LAB) starter cultures based on their technological properties. Seven LAB strains isolated from Tella were characterized for their carbohydrate utilization, salt content, temperature, and acid tolerances, growth and acidification rates, and metabolite profiles. Most strains efficiently utilized various carbohydrates, with Lactiplantibacillus plantarum TDM41 showing exceptional versatility. The strains exhibited similar growth characteristics. Principal component analysis of stress tolerance properties revealed that L. plantarum TDM41, Pediococcus pentosaceus TAA01, and Leuconostoc mesenteroides TDB22 exhibited superior tolerance ability. Strong acidification properties were detected in the L. plantarum TDM41, P. pentosaceus TAA01, and Leuconostoc mesenteroides TDB22 strains after 24 h incubation at 30°C. L. plantarum TDM41 displayed the fastest acidification rate throughout the analysis period. All LAB strains produced significant amounts of diverse organic acids, including lactic acid, citric acid, acetic acid, malic acid, and succinic acid, with lactic acid being the primary acid produced by each strain. Overall, strains L. plantarum TDM41 and P. pentosaceus TAA01 prove to be potential candidates for Tella industrial starter cultures and similar cereal products owing to their robust technological properties.
Introduction
Materials and Methods
Results and Discussion
Conclusion
Acknowledgments
This work was supported by Korea Institute of Planning and Evaluation for Technology in Food, Agriculture and Forestry (IPET) through High Value-added Food Technology Development Program, funded by Ministry of Agriculture, Food and Rural Affairs (MAFRA)(RS-2022-IP322014).
Author Contributions
Gashaw Assefa Yehuala: conceptualization, methodology, investigation, data curation, formal analysis, visualization, writing–original draft. Jaein Choe: reviewing and editing. Nurelegne Tefera Shibeshi: conceptualization, reviewing and editing, and supervision. Kumsa Delessa: conceptualization, reviewing and editing. Asnake Desalegn: conceptualization, reviewing and editing. Mi-Kyung Park: conceptualization, reviewing and editing, and supervision.
Conflict of Interest
The authors have no conflict of interest to report.
Data Availability
All the data related to this study has been presented in the article.
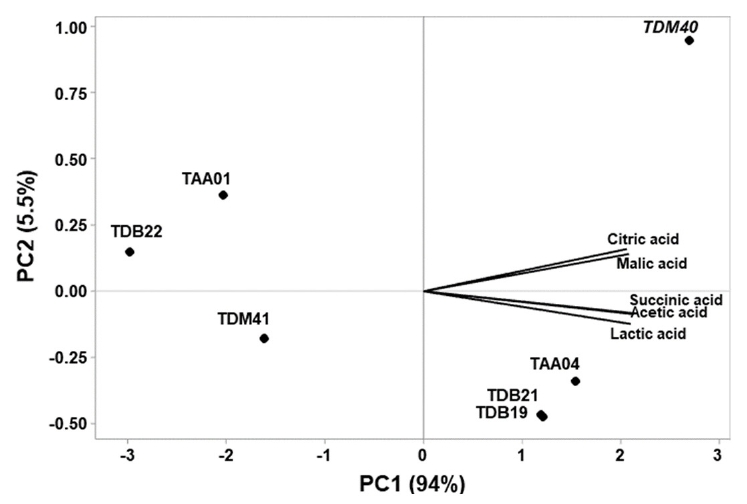
T1, sterile Tella substrate extract (STSE); T2−T8, STSE samples fermented with respective strains; ND, not detected. Values are expressed as the mean of triplicate experiments ± SD. Different lowercase letters in a column indicate statistically significant differences (P < 0.05) among the organic acid contents of the STSE samples fermented with different strains
- Achi OK, Ukwuru M. 2015. Cereal-based fermented foods of Africa as functional foods. Int J Microbiol App. 2(4): 71–83.Article
- Alemayehu HG. 2018. Physico-chemical characterization of commercial local alcohol beverages available in South Nations, Nationalities and People’s Regional State, Ethiopia. Int J ChemTech Res. 11(8): 227–231.Article
- Andualem B, Shiferaw M, Berhane N. 2017. Isolation and characterization of Saccaromyces cervisiae yeasts isolates from “Tella” for beer production. Ann Res Rev Biol. 15(5): 1–12.Article
- Ashenafi M. 2006. A review on the microbiology of indigenous fermented foods and beverages of Ethiopia. Ethiop J Biol Sci. 5(2): 189–245.Article
- Berhanu A. 2014. Microbial profile of Tella and the role of gesho (Rhamnus prinoides) as bittering and antimicrobial agent in traditional Tella (Beer) production. Int Food Res J. 21(1): 357–365.PDF
- Bevilacqua A, Altieri C, Corbo MR, Sinigaglia M, Ouoba LII. 2010. Characterization of lactic acid bacteria isolated from Italian Bella di Cerignola table olives: selection of potential multifunctional starter cultures. J Food Sci. 75(8): M536–M544.ArticlePubMed
- Birhanu AM, Teferra TF, Lema TB. 2021. Fermentation dynamics of Ethiopian traditional beer (Tella) as influenced by substitution of Gesho (Rhamnus prinoides) with Moringa stenopetala: An innovation for nutrition. Int J Food Sci. 2021: 7083638.ArticlePubMedPMCPDF
- Blandino A, Al-Aseeri M, Pandiella S, Cantero D, Webb C. 2003. Cereal-based fermented foods and beverages. Food Res Int. 36(6): 527–543.Article
- Capozzi V, Fiocco D, Spano G. 2011. Responses of lactic acid bacteria to cold stress. In Tsakalidou E, Papadimitriou K. (eds.), Stress Responses of Lactic Acid Bacteria, pp. 91–110, Springer, USA.PDF
- Carnevali P, Ciati R, Leporati A, Paese M. 2007. Liquid sourdough fermentation: Industrial application perspectives. Food Microbiol. 24(2): 150–154.ArticlePubMed
- Celik H, Asik BB, Gurel S, Katkat AV. 2010. Effects of potassium and iron on macro element uptake of maize. Zemdirbyste-Agriculture. 97(1): 11–22.PDF
- Chan MZA, Chua JY, Toh M, Liu SQ. 2019. Survival of probiotic strain Lactobacillus paracasei L26 during co-fermentation with Saccaromyces cervisiae for the development of a novel beer beverage. Food Microbiol. 82: 541–550.ArticlePubMed
- Chen C, Lu Y, Yu H, Chen Z, Tian H. 2019. Influence of 4 lactic acid bacteria on the flavor profile of fermented apple juice. Food Biosci. 27: 30–36.Article
- Coda R, Nionelli L, Rizzello CG, De Angelis M, Tossut P, et al. 2010. Spelt and emmer flours: characterization of the lactic acid bacteria microbiota and selection of mixed starters for bread making. J Appl Microbiol. 108(3): 925–935.ArticlePubMedPDF
- De Angelis M, Gobbetti M. 2004. Environmental stress responses in Lactobacillus: a review. Proteomics. 4(1): 106–122.ArticlePubMedPDF
- Di Cagno R, Coda R, De Angelis M, Gobbetti M. 2013. Exploitation of vegetables and fruits through lactic acid fermentation. Food Microbiol. 33(1): 1–10.ArticlePubMed
- Divyashree S, Ramu R, Sreenivasa MY. 2024. Evaluation of new candidate probiotic Lactobacillus strains isolated from a traditional fermented food-multigrain-millet dosa batter. Food Biosci. 57: 103450.Article
- dos Santos DC, de Oliveira Filho JG, Andretta JR, Silva FG, Egea MB. 2023. Challenges in maintaining the probiotic potential in alcoholic beverage development. Food Biosci. 52: 102485.Article
- Fentie EG, Emire SA, Demsash HD, Dadi DW, Shin JH. 2020. Cereal-and fruit-based Ethiopian traditional fermented alcoholic beverages. Foods. 9(12): 1781.ArticlePubMedPMC
- Ferreira I, de Sousa Melo D, Menezes AGT, Fonseca HC, de Assis BBT, et al. 2022. Evaluation of potentially probiotic yeasts and Lactiplantibacillus plantarum in co-culture for the elaboration of a functional plant-based fermented beverage. Food Res Int. 160: 111697.ArticlePubMed
- Gänzle MG. 2014. Enzymatic and bacterial conversions during sourdough fermentation. Food Microbiol. 37(2): 10.Article
- Gobbetti M. 1998. The sourdough microflora: Interactions of lactic acid bacteria and yeasts. Trends Food Sci Technol. 9(7): 267–274.Article
- Goswami G, Bora SS, Parveen A, Boro RC, Barooah M. 2017. Identification and functional properties of dominant lactic acid bacteria isolated from Kahudi, a traditional rapeseed fermented food product of Assam, India. J Ethn Foods. 4(3): 187–197.Article
- Guan N, Liu L. 2020. Microbial response to acid stress: mechanisms and applications. Appl Microbiol Biotechnol. 104(1): 51–65.ArticlePubMedPMCPDF
- Guetarni H. 2024. Biochemical, Physiological and Technological Properties of Lactic Acid Bacteria Isolated and Characterized from Hard and Soft Wheat: Lactic acid bacteria isolated and characterized from wheat. Ann Arid Zone. 63(1): 9–21.Article
- Hedberg M, Hasslöf P, Sjöström I, Twetman S, Stecksén‐Blicks C. 2008. Sugar fermentation in probiotic bacteria-an in vitro study. Oral Microbiol Immunol. 23(6): 482–485.ArticlePubMed
- Hinojosa-Avila CR, García-Gamboa R, Chedraui-Urrea JJ, García-Cayuela T. 2023. Exploring the potential of probiotic-enriched beer: Microorganisms, fermentation strategies, sensory attributes, and health implications. Food Res Int. 175: 113717.ArticlePubMed
- Holzapfel WH. 2002. Appropriate starter culture technologies for small-scale fermentation in developing countries. Int J Food Microbiol. 75(3): 197–212.ArticlePubMed
- Hotessa N, Robe J. 2020. Ethiopian indigenous traditional fermented beverage: the role of the microorganisms toward nutritional and safety value of fermented beverage. Int J Microbiol. 2020: 8891259.ArticlePubMedPMCPDF
- Hu Y, Zhang L, Badar IH, Liu Q, Liu H, et al. 2024. Insights into the flavor perception and enhancement of sodium-reduced fermented foods: A review. Crit Rev Food Sci Nutr. 64(8): 2248–2262.ArticlePubMed
- Kang O, Vézinz LP, Laberge S, Simard R. 1998. Some factors influencing the autolysis of Lactobacillus bulgaricus and Lactobacillus casei. J Dairy Sci. 81: 639–646.Article
- Kang W, Pan L, Peng C, Dong L, Cao S, et al. 2020. Isolation and characterization of lactic acid bacteria from human milk. J Dairy Sci. 103: 9980–9991.ArticlePubMed
- Kathade SA, Aswani MA, Anand PK, Jagtap S, Bipinraj NK. 2020. Isolation of Lactobacillus from donkey dung and its probiotic characterization. Korean J Microbiol. 56(2): 160–169.PDF
- Lee M, Regu M, Seleshe S. 2015. Uniqueness of Ethiopian traditional alcoholic beverage of plant origin, Tella. J Ethn Foods. 2(3): 110–114.Article
- Lemi BW. 2020. Microbiology of Ethiopian traditionally fermented beverages and condiments. Int J Microbiol. 2020: 1478536.ArticlePubMedPMC
- Manini F, Casiraghi M, Poutanen K, Brasca M, Erba D, et al. 2016. Characterization of lactic acid bacteria isolated from wheat bran sourdough. LWT-Food Sci Technol. 66: 275–283.Article
- Marklinder I, Lönner C. 1992. Fermentation properties of intestinal strains of Lactobacillus, of a sour dough and of a yoghurt starter culture in an oat-based nutritive solution. Food Microbiol. 9(3): 197–205.Article
- Massera A, Assof M, Sari S, Ciklic I, Mercado L, et al. 2021. Effect of low temperature fermentation on the yeast-derived volatile aroma composition and sensory profile in Merlot wines. LWT-Food Sci Technol. 142: 111069.Article
- Montemurro M, Celano G, DeAngelis M, Gobbetti M, Rizzello CG, et al. 2020. Selection of non-Lactobacillus strains to be used as starters for sourdough fermentation. Food Microbiol. 90: 103491.ArticlePubMed
- Nout MJR, Sarkar PK. 1999. Lactic acid food fermentation in tropical climates. In Lactic Acid Bacteria: Genetics, Metabolism and Applications: Proceedings of the Sixth Symposium on lactic acid bacteria: genetics, metabolism and applications. 19-23 September 1999. Veldhoven, The Netherlands: 395–401. Springer Netherlands.Article
- Nuryana I, Andriani A, Lisdiyanti P, Yopi. 2019. Analysis of organic acids produced by lactic acid bacteria. In IOP Conference Series: Earth and Environmental Science, Vol. 251, p. 012054, IOP Publishing.PDF
- Puerari C, Magalhães-Guedes KT, Schwan RF. 2015. Physicochemical and microbiological characterization of chicha, a rice-based fermented beverage produced by Umutina Brazilian Amerindians. Food Microbiol. 46: 210–217.ArticlePubMed
- Rahmati F. 2017. Characterization of Lactobacillus, Bacillus and Saccharomyces isolated from Iranian traditional dairy products for potential sources of starter cultures. AIMS Microbiol. 3(4): 815–825.ArticlePubMedPMC
- Sáez GD, Hébert EM, Saavedra L, Zárate G. 2017. Molecular identification and technological characterization of lactic acid bacteria isolated from fermented kidney beans flours (Phaseolus vulgaris L. and P. coccineus) in northwestern Argentina. Food Res Int. 102: 605–615.ArticlePubMed
- Salvucci E, LeBlanc JG, Pérez G. 2016. Technological properties of Lactic acid bacteria isolated from raw cereal material. LWT-Food Sci Technol. 70: 185–191.Article
- Samuel S, Berhanu A. 1991. The microbiology of TELLA fermentation. Sinet. 14: 81–92.
- Sánchez García B, Champomier-Vergès MC, Collado MC, Anglade P, Baraige F, et al. 2007. Low pH adaptation and the acid tolerance response of Bifidobacterium longum biotype longum. Appl Environ Microbiol. 73(20): 6450–6459.ArticlePubMedPMCPDF
- Sawadogo‐Lingani H, Diawara B, Traore A, Jakobsen M. 2008. Technological properties of Lactobacillus fermentum involved in the processing of dolo and pito, West African sorghum beers, for the selection of starter cultures. J Appl Microbiol. 104(3): 873–882.ArticlePubMed
- Shewakena S, Chandravanshi B, Debebe A. 2017. Levels of total polyphenol, flavonoid, tannin and antioxidant activity of selected Ethiopian fermented traditional beverages. Int Food Res J. 24(5): 2033–2040.PDF
- Smid EJ, Lacroix C. 2013. Microbe-microbe interactions in mixed culture food fermentations. Curr Opin Biotechnol. 24(2): 148–154.ArticlePubMed
- Speranza B, Racioppo A, Beneduce L, Bevilacqua A, Sinigaglia M, et al. 2017. Autochthonous lactic acid bacteria with probiotic aptitudes as starter cultures for fish-based products. Food Microbiol. 65: 244–253.ArticlePubMed
- Tamene A, Baye K, Kariluoto S, Edelmann M, Bationo F, et al. 2019. Lactobacillus plantarum P2R3FA isolated from traditional cereal-based fermented food increase folate status in deficient rats. Nutrients. 11(11): 2819.ArticlePubMedPMC
- Tangyu M, Muller J, Bolten CJ, Wittmann C. 2019. Fermentation of plant-based milk alternatives for improved flavour and nutritional value. Appl Microbiol Biotechnol. 103(23-24): 9263–9275.ArticlePubMedPMCPDF
- Tekle B, Anuradha Jabasingh S, Fantaw D, Gebreslassie T, Ram Mohan Rao S, et al. 2019. An insight into the Ethiopian traditional alcoholic beverage: Tella processing, fermentation kinetics, microbial profiling and nutrient analysis. LWT-Food Sci Technol. 107: 9–15.Article
- Üçok G, Sert D. 2020. Growth kinetics and biomass characteristics of Lactobacillus plantarum L14 isolated from sourdough: Effect of fermentation time on dough machinability. LWT-Food Sci Technol. 129: 109516.Article
- Van De Guchte M, Serror P, Chervaux C, Smokvina T, Ehrlich SD, et al. 2002. Stress responses in lactic acid bacteria. In Lactic Acid Bacteria: Genetics, Metabolism and Applications: Proceedings of the seventh Symposium on lactic acid bacteria: genetics, metabolism and applications. 1-5 September 2002. Egmond aan Zee, the Netherlands, pp. 187-216, Springer Netherlands.PDF
- Vermeiren L, Devlieghere F, Debevere J. 2004. Evaluation of meat born lactic acid bacteria as protective cultures for the biopreservation of cooked meat products. Int J Food Microbiol. 96(2): 149–164.ArticlePubMed
- Vinicius De Melo Pereira G, De Carvalho Neto DP, Junqueira ACDO, Karp SG, Letti LA, et al. 2020. A review of selection criteria for starter culture development in the food fermentation industry. Food Rev Int. 36(2): 135–167.Article
- Wang C, Song X, Li C, He L, Wang X, et al. 2023. Mixed fermentation with Lactobacillus plantarum, Bifidobacterium animalis subsp. lactis and Candida utilis improves the fermentation quality of Hong Suan Tang. Food Chem. 402: 134488.ArticlePubMed
- Wu C, Zhang J, Chen W, Wang M, Du G, et al. 2012. A combined physiological and proteomic approach to reveal lactic-acid-induced alterations in Lactobacillus casei Zhang and its mutant with enhanced lactic acid tolerance. Appl Microbiol Biotechnol. 93(2): 707–722.ArticlePubMedPDF
- Xiong T, Li X, Guan Q, Peng F, Xie M. 2014. Starter culture fermentation of Chinese sauerkraut: Growth, acidification and metabolic analyses. Food Control. 41: 122–127.Article
- Yehuala GA, Shibeshi NT, Kim SH, Park MK. 2024. Characterization of autochthonous lactic acid bacteria isolated from a traditional Ethiopian beverage, Tella. Foods. 13(4): 575.ArticlePubMedPMC
- Yohannes T, Melak F, Siraj K. 2013. Preparation and physicochemical analysis of some Ethiopian traditional alcoholic beverages. African J Food Sci. 7(11): 399–403.Article
- Yousef AE, Courtney PD. 2003. Basics of stress adaptation and implications in new-generation foods. In Microbial stress adaptation and food safety, 1st ed., pp. 1–30, Taylor and Francis Group, New York.PDF
- Zvauya R, Mygochi T, Parawira W. 1997. Microbial and biochemical changes occurring during production of masvusvu and mangisi, traditional Zimbabwean beverages. Plant Foods Hum Nutr. 51(1): 43–51.ArticlePubMed
References
Figure & Data
References
Citations

- Preparation method and physicochemical characteristics of Tella: an Ethiopian fermented beverage
Rabira Lemessa Gudeta, Solomon Abera, Hirpha Adugna Areti
Journal of Ethnic Foods.2025;[Epub] CrossRef
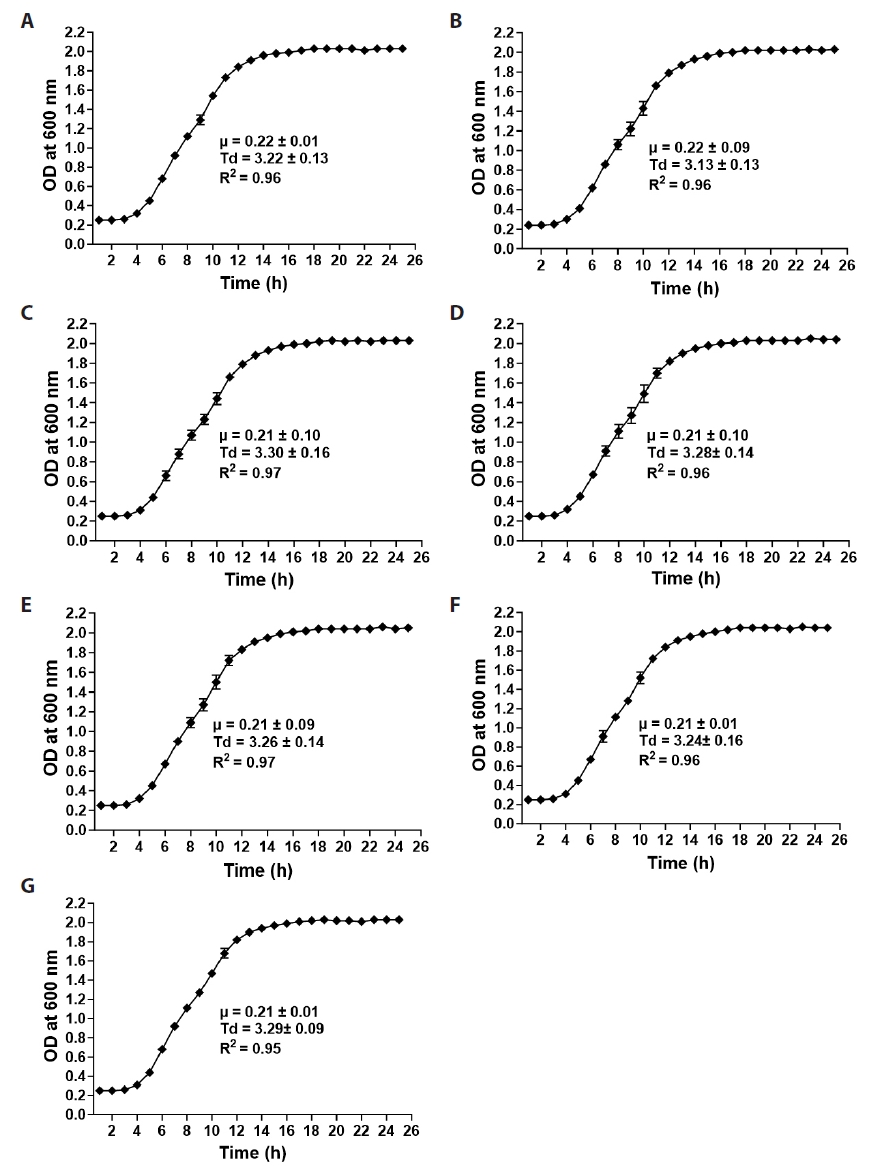
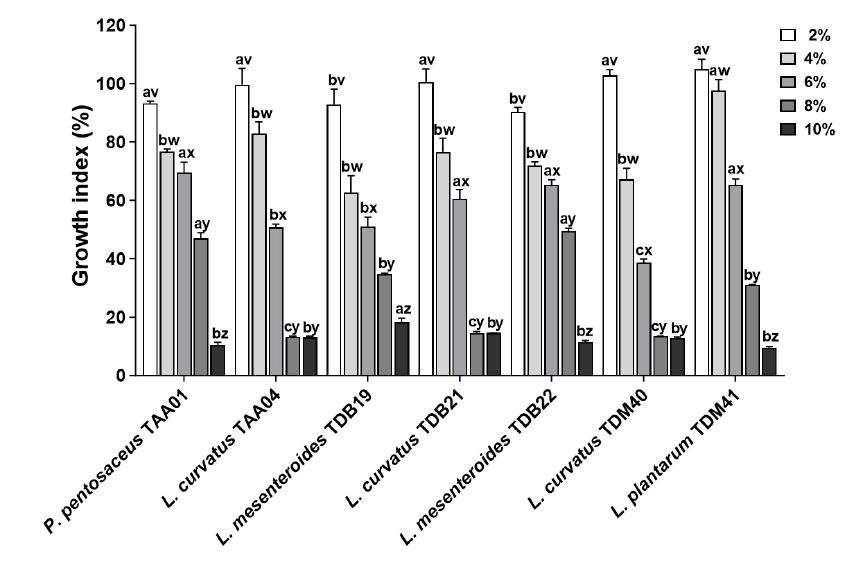
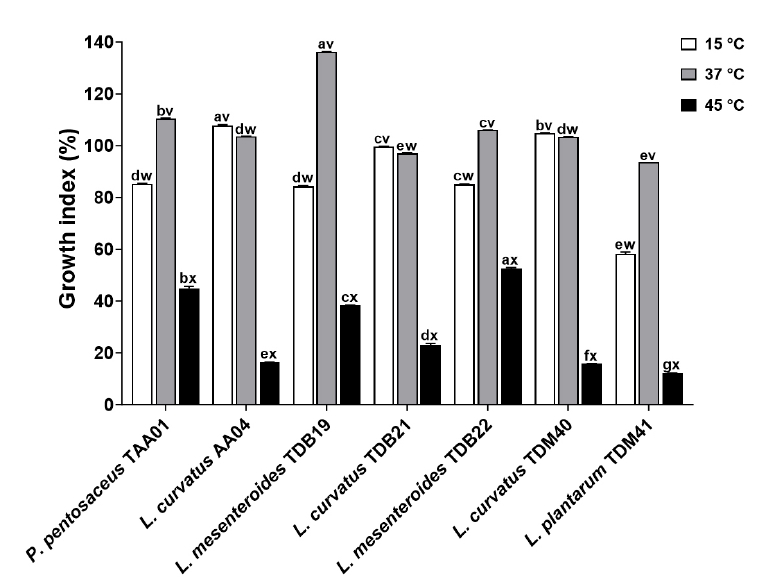
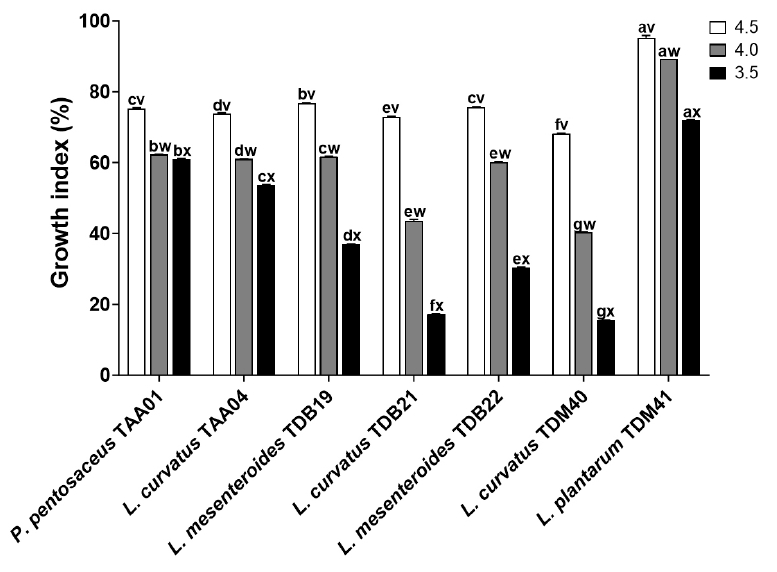
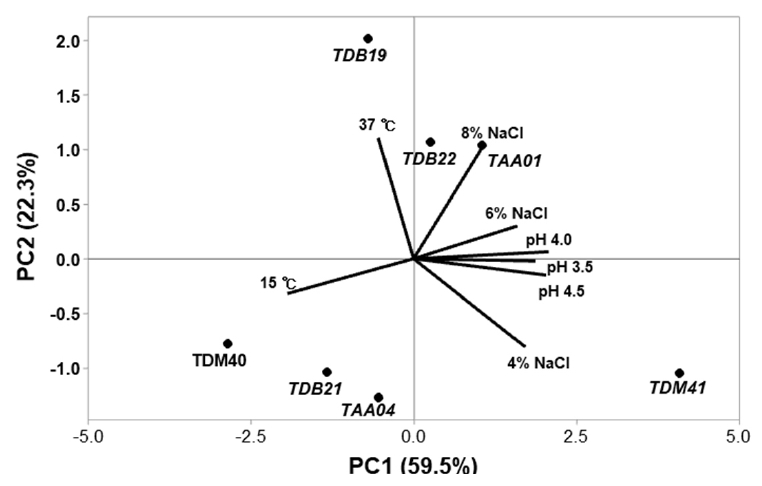
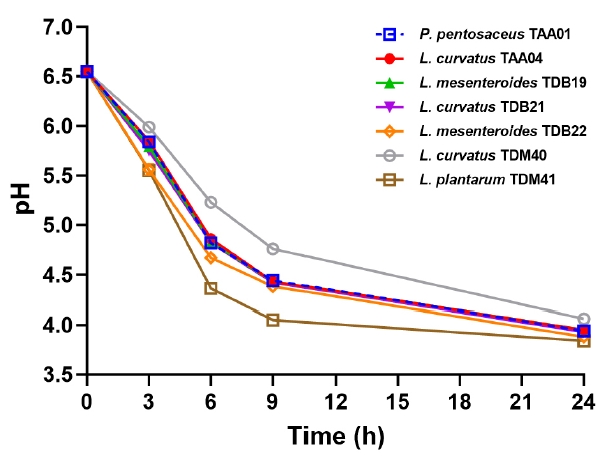
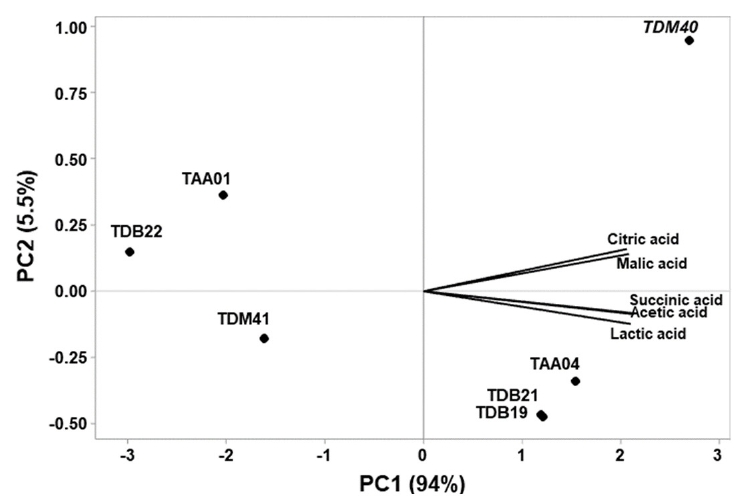
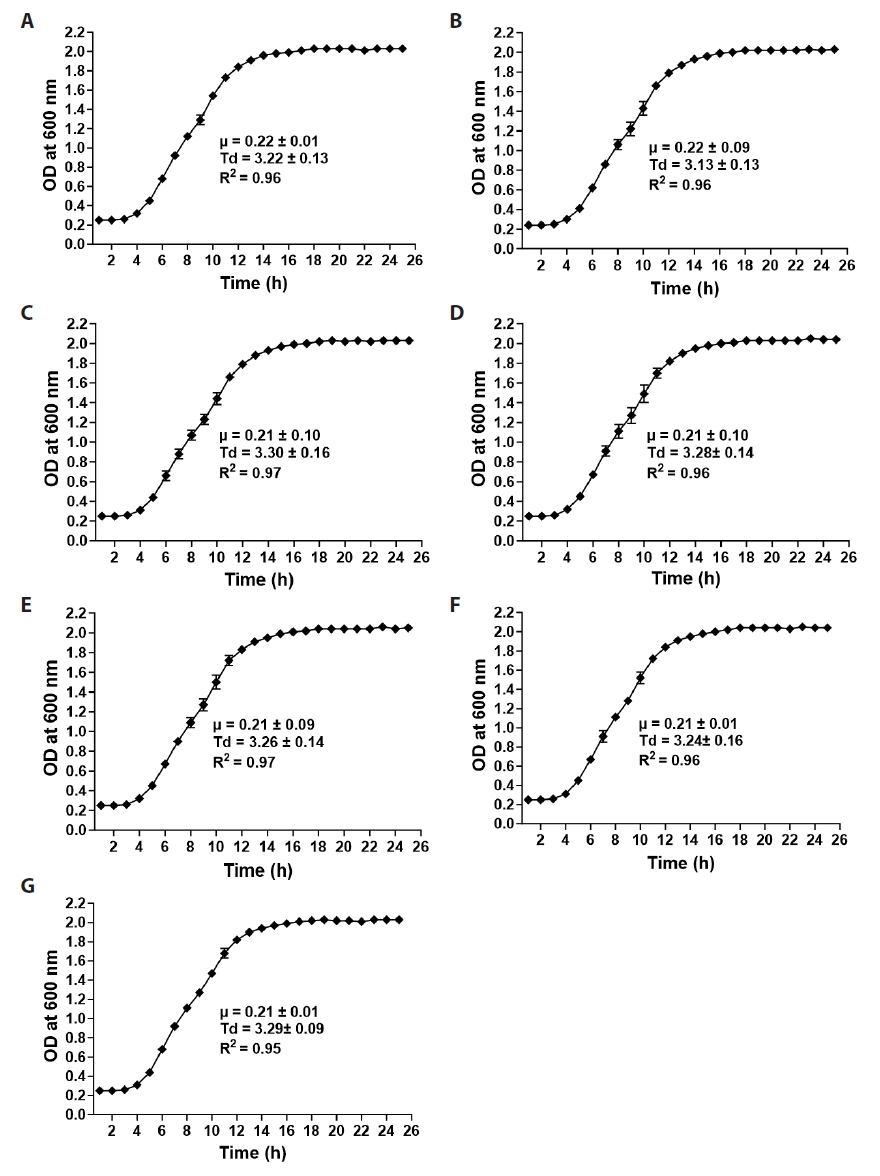
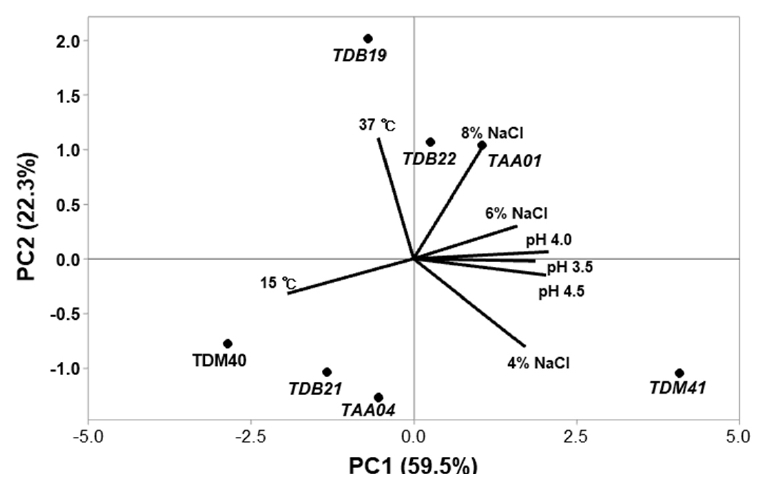
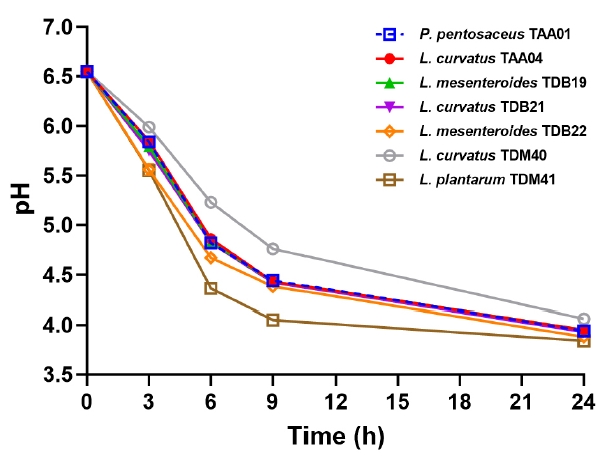
Fig. 1.
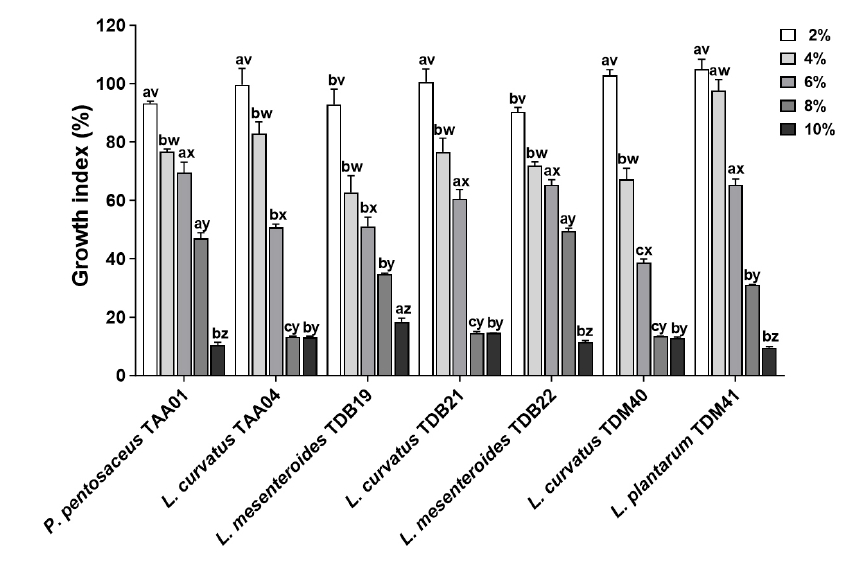
Fig. 2.
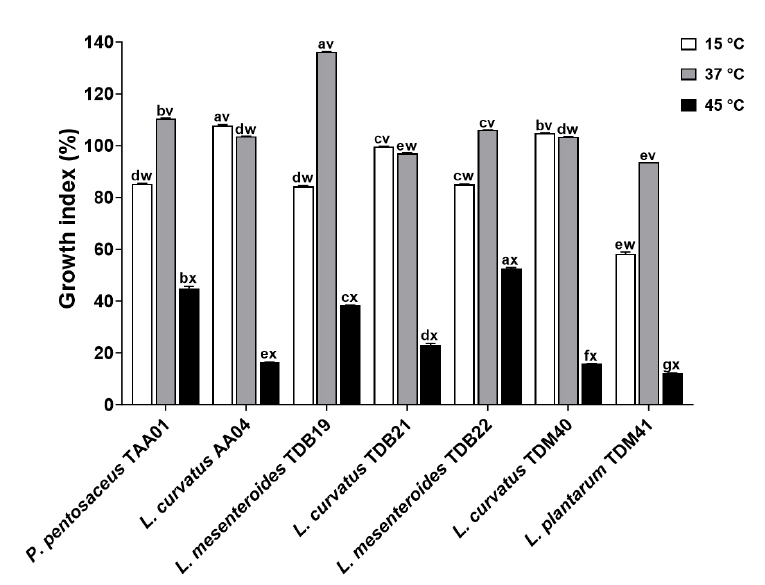
Fig. 3.
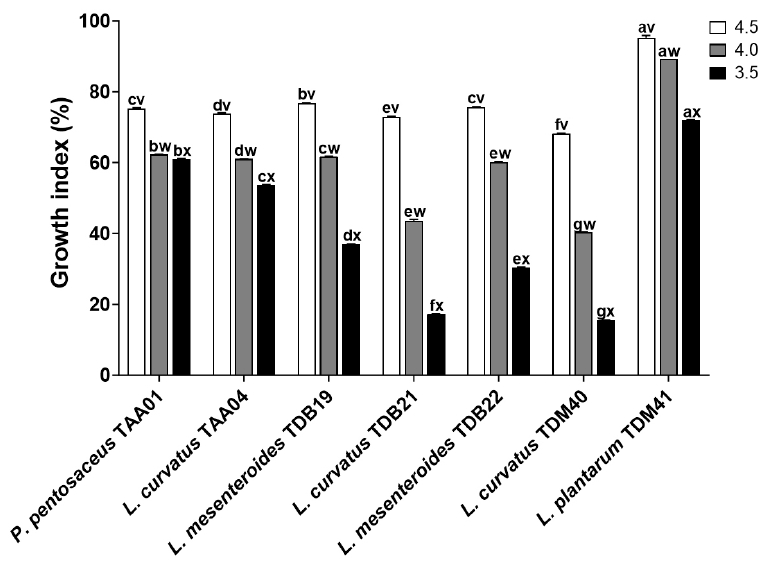
Fig. 4.
Fig. 5.
Fig. 6.
Fig. 7.
| LAB strains | Carbohydrate utilization |
|||||
|---|---|---|---|---|---|---|
| Glucose | Sucrose | Maltose | Fructose | Galactose | Sorbitol | |
| P. pentosaceus TAA01 | ++ | ++ | ++ | ++ | ++ | – |
| L. curvatus TAA04 | ++ | ++ | ++ | ++ | ++ | – |
| L. mesenteroides TDB19 | ++ | ++ | ++ | ++ | ++ | – |
| L. curvatus TDB21 | ++ | ++ | ++ | ++ | ++ | + |
| L. mesenteroides TDB22 | ++ | ++ | ++ | ++ | ++ | – |
| L. curvatus TDM40 | ++ | ++ | ++ | ++ | ++ | – |
| L. plantarum TDM41 | +++ | +++ | +++ | +++ | +++ | + |
| Samples | Organic acid concentrations (g/L) |
||||
|---|---|---|---|---|---|
| Lactic acid | Acetic acid | Citric acid | Malic acid | Succinic acid | |
| T1 | 0.02 ± 0.00e | ND | 0.03 ± 0.00f | 0.02 ± 0.00e | ND |
| T2 | 1.09 ± 0.04c | 0.08 ± 0.00b | 0.72 ± 0.04c | 0.81 ± 0.05c | 0.07 ± 0.00a |
| T3 | 2.12 ± 0.11a | 0.25 ± 0.01a | 1.07 ± 0.05b | 1.53 ± 0.08b | 0.14 ± 0.01a |
| T4 | 2.13 ± 0.11a | 0.24 ± 0.01a | 0.99 ± 0.05b | 1.43 ± 0.07b | 0.13 ± 0.01a |
| T5 | 2.15 ± 0.11a | 0.24 ± 0.01a | 0.99 ± 0.05b | 1.44 ± 0.07b | 0.13 ± 0.01a |
| T6 | 0.85 ± 0.04d | 0.07 ± 0.00b | 0.48 ± 0.02e | 0.62 ± 0.03d | 0.05 ± 0.00a |
| T7 | 2.15 ± 0.11a | 0.26 ± 0.02a | 1.47 ± 0.07a | 2.22 ± 0.13a | 0.14 ± 0.01a |
| T8 | 1.38 ± 0.06b | 0.11 ± 0.01b | 0.65 ± 0.03d | 0.81 ± 0.04c | 0.08 ± 0.00a |
+++, high utilization (ΔpH ≥ 3.5); ++, medium utilization (2.0 ≤ ΔpH < 3.5); +, low utilization (0.5 ≤ ΔpH < 2.0); –, no utilization (ΔpH < 0.5) after 48-h incubation at 30°C
T1, sterile
Table 1.
Table 2.
TOP
 MSK
MSK

 ePub Link
ePub Link Cite this Article
Cite this Article








