Articles
- Page Path
- HOME > J. Microbiol > Volume 63(2); 2025 > Article
-
Full article
LasB activation in Pseudomonas aeruginosa: Quorum sensing-mediated release of an auto-activation inhibitor -
Cheol Seung Lee1, Xi-Hui Li1,2, Chae-Ran Jeon1, Joon-Hee Lee1,2,*

-
Journal of Microbiology 2025;63(2):e2411005.
DOI: https://doi.org/10.71150/jm.2411005
Published online: February 27, 2025
1Department of Pharmacy, College of Pharmacy, Pusan National University, Busan 46241, Republic of Korea
2Research Institute for Drug Development, Pusan National University, Busan 46241, Republic of Korea
- *Correpondence Joon-Hee Lee joonhee@pusan.ac.kr
© The Microbiological Society of Korea
This is an Open Access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/4.0) which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
- 1,744 Views
- 81 Download
- 1 Scopus
ABSTRACT
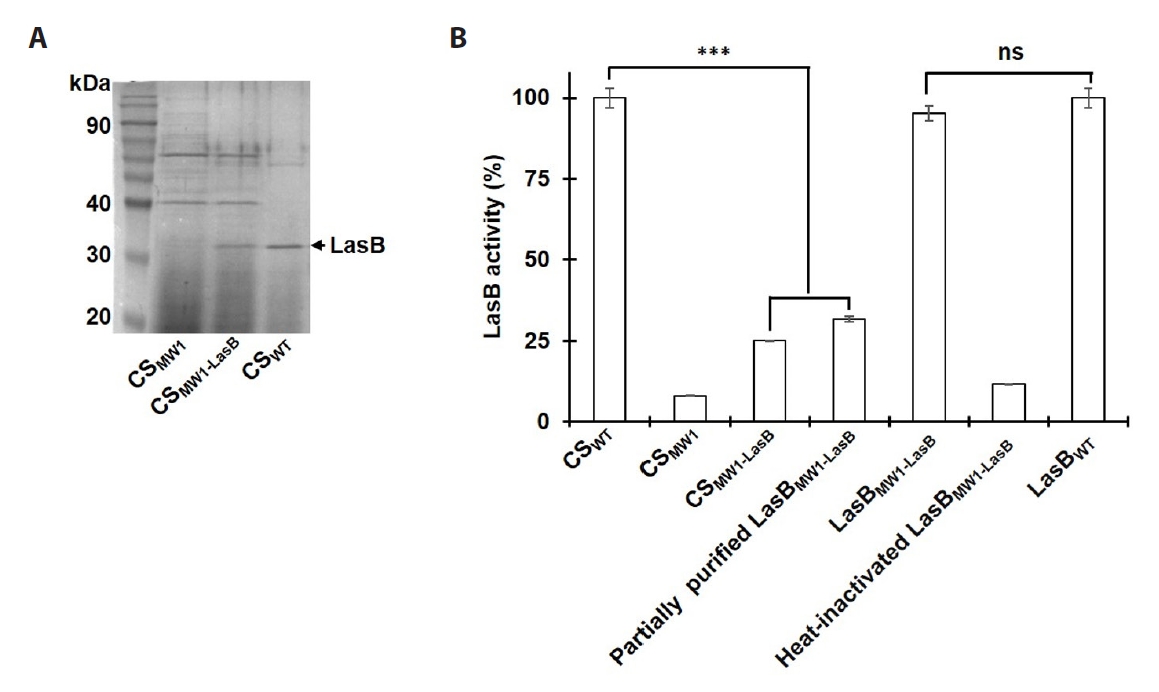
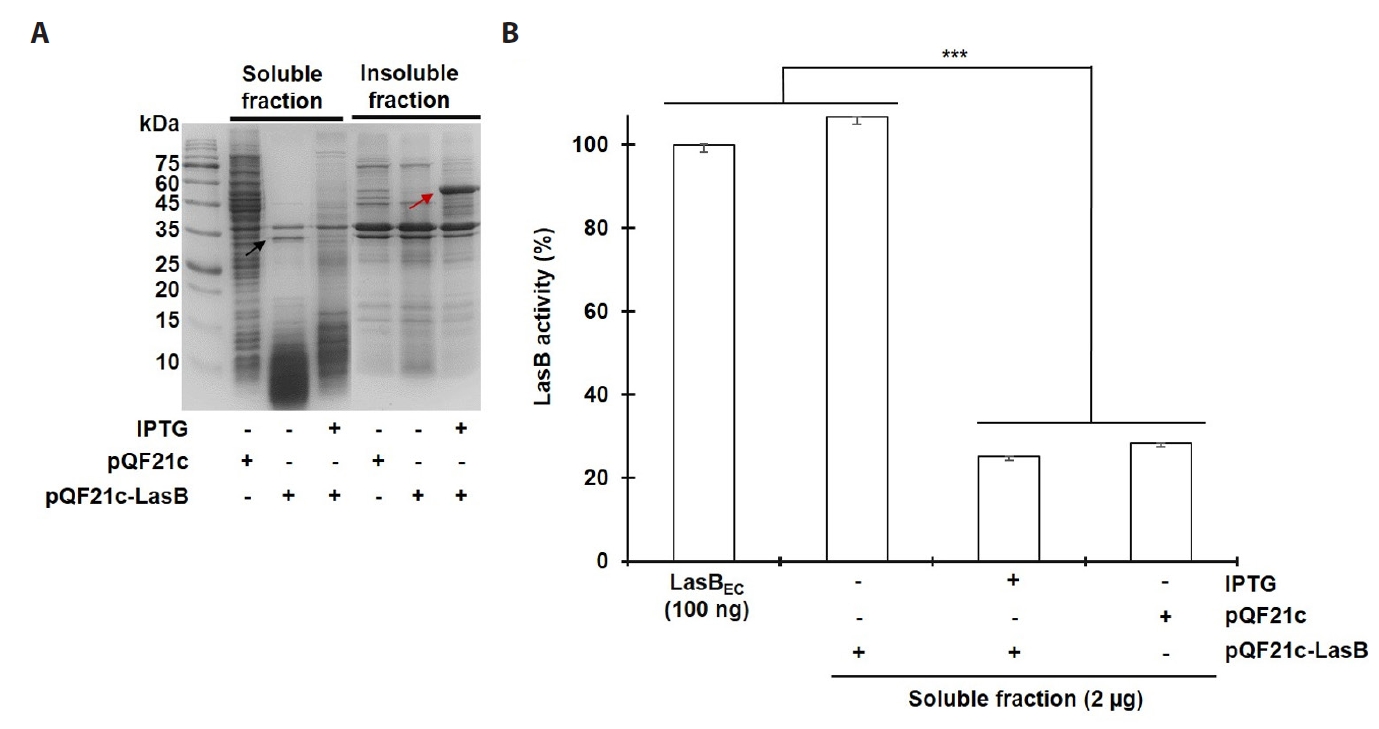
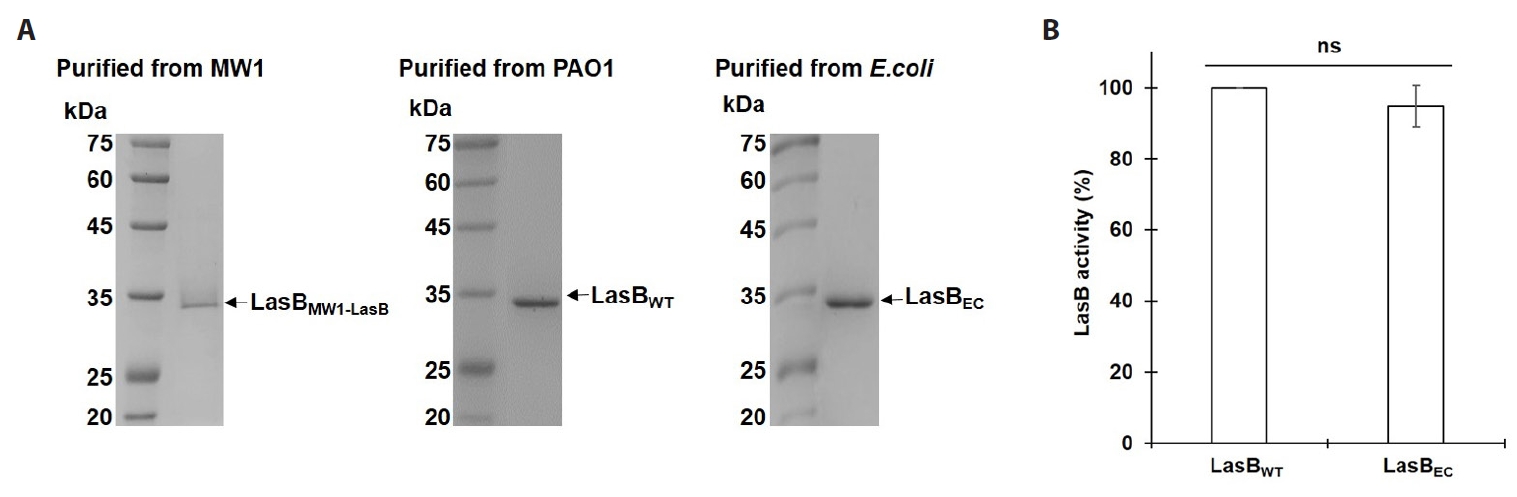
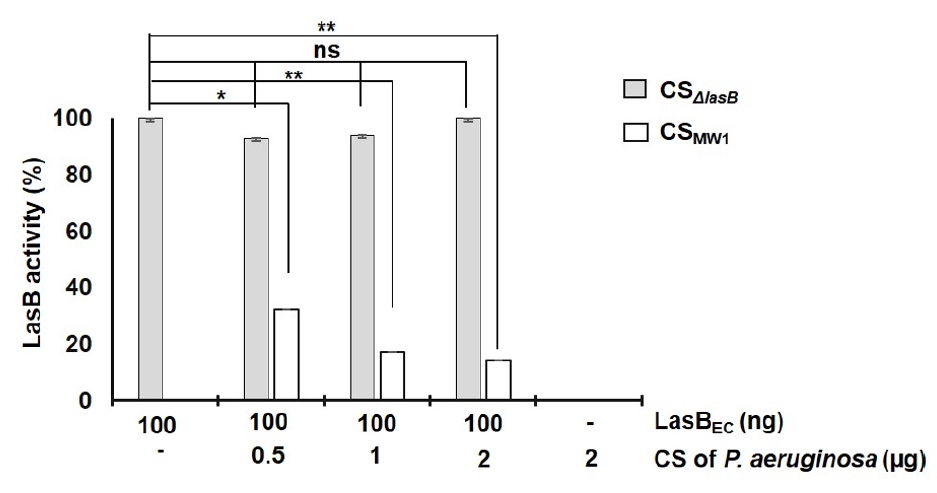
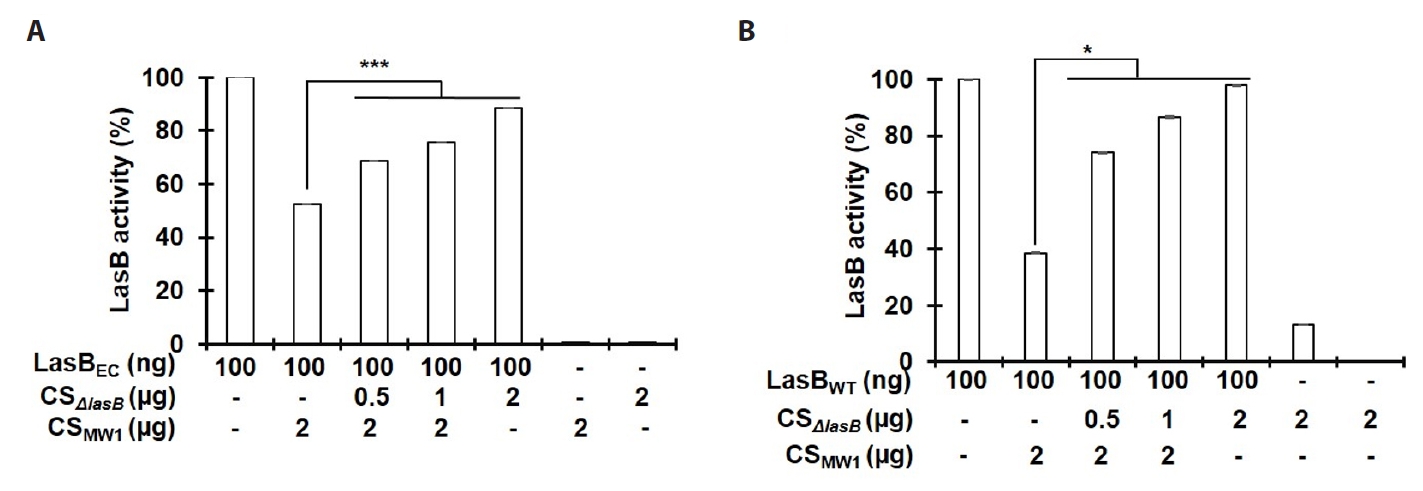
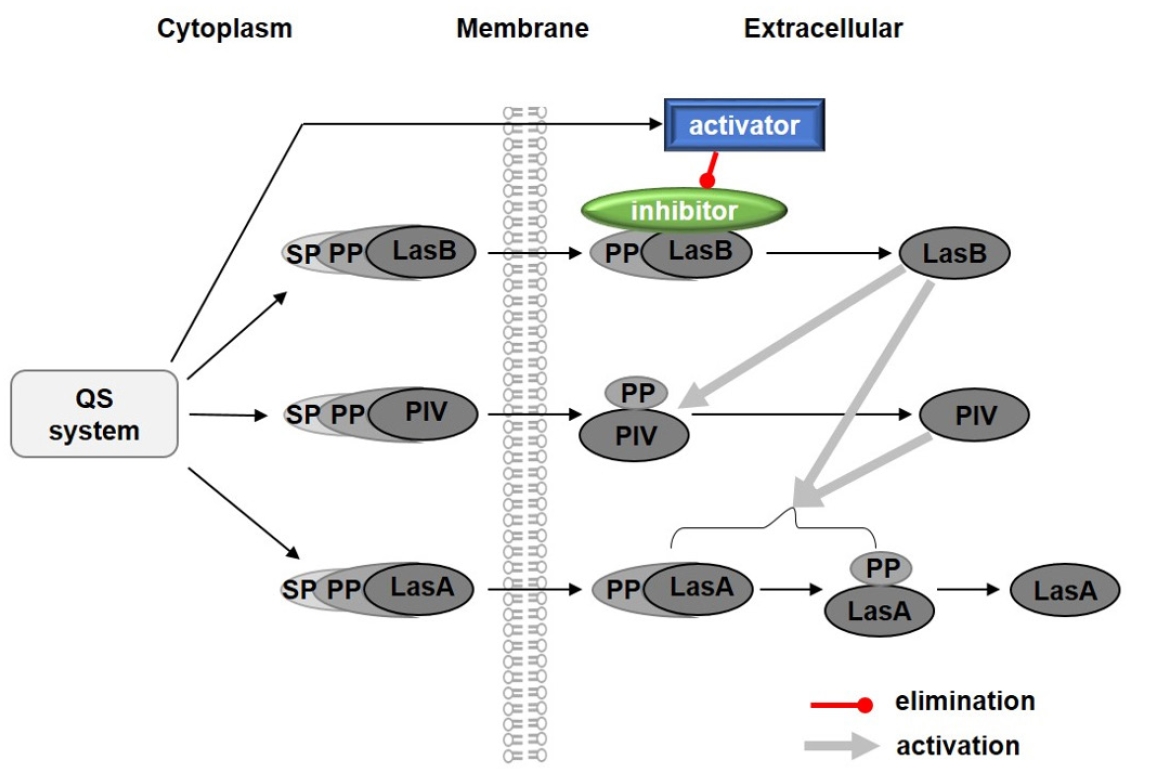
- Pseudomonas aeruginosa secretes three major proteases: elastase B (LasB), protease IV (PIV), and elastase A (LasA), which play crucial roles in infection and pathogenesis. These proteases are activated sequentially from LasB in a proteolytic cascade, and LasB was previously thought to undergo auto-activation. However, our previous study suggested that LasB cannot auto-activate independently but requires additional quorum sensing (QS)-dependent factors for activation, as LasB remained inactive in QS-deficient P. aeruginosa (QS-) even under artificial overexpression. In this study, we provide evidence for the existence of a LasB inhibitor in QS- mutants: inactive LasB overexpressed in QS- strains was in its processed form and could be reactivated upon purification; when full-length LasB was overexpressed in Escherichia coli, a heterologous bacterium lacking both LasB activators and inhibitors, the protein underwent normal processing and activation; and purified active LasB was significantly inhibited by culture supernatant (CS) from QS- strains but not by CS from QS+ strains. These findings demonstrate that a LasB inhibitor exists in QS- strains, and in its absence, LasB can undergo auto-activation without requiring an activator. Based on these results, we propose an updated hypothesis: the QS-dependent LasB activator functions by removing the LasB inhibitor rather than acting directly on LasB itself, thus preventing premature LasB activation until QS response is initiated.
Introduction
Materials and Methods
Results
Discussion
Acknowledgments
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (RS-2024-00353202).
Conflict of Interest
We declare that we have no conflicts of interest with the contents of this article.
| Name | Description | References |
|---|---|---|
| Pseudomonas aeruginosa | ||
| PAO1 | Wild type P. aeruginosa, QS+ | (Pearson and Pesci et al., 1997) |
| MW1 | lasI-, rhlI- double mutant of PAO1, TcR, QS- | (Whiteley et al., 1999) |
| ΔlasB | lasB- mutant of PAO1, TcR, QS+ | (Li and Lee, 2019) |
| Escherichia coli | ||
| DH5α | supE44ΔlacU169(80lacZΔM15) hsdR17 recA1 gyrA96thi-1 relA1 | Lab. collection |
| BL21(DE3) | F-ompT hsdSB (rB- mB-) dcm gal λ (DE3) | Lab. collection |
| Plasmids | ||
| pJN105 | araC-pBAD promoter fusion plasmid, GmR | (Newman and Fuqua, 1999) |
| pSP201 | PA3724 (lasB) in pJN105, GmR | (Park et al., 2014) |
| pQF21c | Overexpression plasmid for C-terminal histidine-tagged protein (a modified pET21c with a broad-host-range replication origin, Ori1600), replicable in P. aeruginosa, CbR (ApR for E. coli) | (Park et al., 2014) |
| pQF21c-LasB | lasB in pQF21c, CbR (ApR for E. coli) | (Li and Lee, 2019) |
- Albus AM, Pesci EC, Runyen-Janecky LJ, West SE, Iglewski BH. 1997. Vfr controls quorum sensing in Pseudomonas aeruginosa. J Bacteriol. 179(12): 3928–3935. ArticlePubMedPMCPDF
- Ding F, Oinuma KI, Smalley NE, Schaefer AL, Hamwy O, et al. 2018. The Pseudomonas aeruginosa orphan quorum sensing signal receptor QscR regulates global quorum sensing gene expression by activating a single linked operon. MBio. 9(4): e01274–18. ArticlePubMedPMCPDF
- Horcajada JP, Montero M, Oliver A, Sorlí L, Luque S, et al. 2019. Epidemiology and treatment of multidrug-resistant and extensively drug-resistant Pseudomonas aeruginosa infections. Clin Microbiol Rev. 32(4): e00031–19. ArticlePubMedPMCPDF
- Kessler E, Safrin M. 1988. Partial purification and characterization of an inactive precursor of Pseudomonas aeruginosa elastase. J Bacteriol. 170(3): 1215–1219. ArticlePubMedPMCPDF
- Kessler E, Safrin M, Gustin JK, Ohman DE. 1998. Elastase and the LasA protease of Pseudomonas aeruginosa are secreted with their propeptides. J Biol Chem. 273(46): 30225–30231. ArticlePubMed
- Kostylev M, Kim DY, Smalley NE, Salukhe I, Greenberg EP, et al. 2019. Evolution of the Pseudomonas aeruginosa quorum-sensing hierarchy. Proc Natl Acad Sci USA. 116(14): 7027–7032. ArticlePubMedPMC
- Li XH, Lee JH. 2019. Quorum sensing-dependent post-secretional activation of extracellular proteases in Pseudomonas aeruginosa. J Biol Chem. 294(51): 19635–19644. ArticlePubMedPMC
- McIver K, Kessler E, Ohman DE. 1991. Substitution of active-site His-223 in Pseudomonas aeruginosa elastase and expression of the mutated lasB alleles in Escherichia coli show evidence for autoproteolytic processing of proelastase. J Bacteriol. 173(24): 7781–7789. ArticlePubMedPMCPDF
- McIver KS, Olson JC, Ohman DE. 1993. Pseudomonas aeruginosa lasB1 mutants produce an elastase, substituted at active-site His-223, that is defective in activity, processing, and secretion. J. Bacteriol. 175(13): 4008–4015. ArticlePubMedPMCPDF
- McIver KS, Kessler E, Olson JC, Ohman DE. 1995. The elastase propeptide functions as an intramolecular chaperone required for elastase activity and secretion in Pseudomonas aeruginosa. Mol Microbiol. 18(5): 877–889. ArticlePubMed
- Miller MB, Bassler BL. 2001. Quorum sensing in bacteria. Annu Rev Microbiol. 55(1): 165–199. ArticlePubMed
- Miranda SW, Asfahl KL, Dandekar AA, Greenberg EP. 2022. Pseudomonas aeruginosa quorum sensing. Adv Exp Med Biol. 1386: 95–115. ArticlePubMedPMC
- Moradali MF, Ghods S, Rehm BH. 2017. Pseudomonas aeruginosa lifestyle: a paradigm for adaptation, survival, and persistence. Front Cell Infect Microbiol. 7: 39.ArticlePubMedPMC
- Newman JR, Fuqua C. 1999. Broad-host-range expression vectors that carry the L-arabinose-inducible Escherichia coli araBAD promoter and the araC regulator. Gene. 227(2): 197–203. ArticlePubMed
- Park SJ, Kim SK, So YI, Park HY, Li XH, et al. 2014. Protease IV, a quorum sensing‐dependent protease of Pseudomonas aeruginosa modulates insect innate immunity. Mol Microbiol. 94(6): 1298–1314. ArticlePubMed
- Pearson JP, Pesci EC, Iglewski BH. 1997. Roles of Pseudomonas aeruginosa las and rhl quorum-sensing systems in control of elastase and rhamnolipid biosynthesis genes. J. Bacteriol. 179(18): 5756–5767. ArticlePubMedPMCPDF
- Qin S, Xiao W, Zhou C, Pu Q, Deng X, et al. 2022. Pseudomonas aeruginosa: pathogenesis, virulence factors, antibiotic resistance, interaction with host, technology advances and emerging therapeutics. Signal Transduct Target Ther. 7(1): 1–27. ArticlePubMedPMCPDF
- Shin Y, Li XH, Lee CS, Lee JH. 2022. Mutational analysis on stable expression and LasB inhibition of LasB propeptide in Pseudomonas aeruginosa. J Microbiol. 60(7): 727–734.ArticlePubMedPDF
- Whiteley M, Lee KM, Greenberg EP. 1999. Identification of genes controlled by quorum sensing in Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 96(24): 13904–13909. ArticlePubMedPMC
- Williams SC, Patterson EK, Carty NL, Griswold JA, Hamood AN, et al. 2004. Pseudomonas aeruginosa autoinducer enters and functions in mammalian cells. J Bacteriol. 186(8): 2281–2287. ArticlePubMedPMCPDF
- Yang D, Hao S, Zhao L, Shi F, Ye G, et al. 2021. Paeonol attenuates quorum-sensing regulated virulence and biofilm formation in Pseudomonas aeruginosa. Front Microbiol. 12: 692474.ArticlePubMedPMC
References
Figure & Data
References
Citations


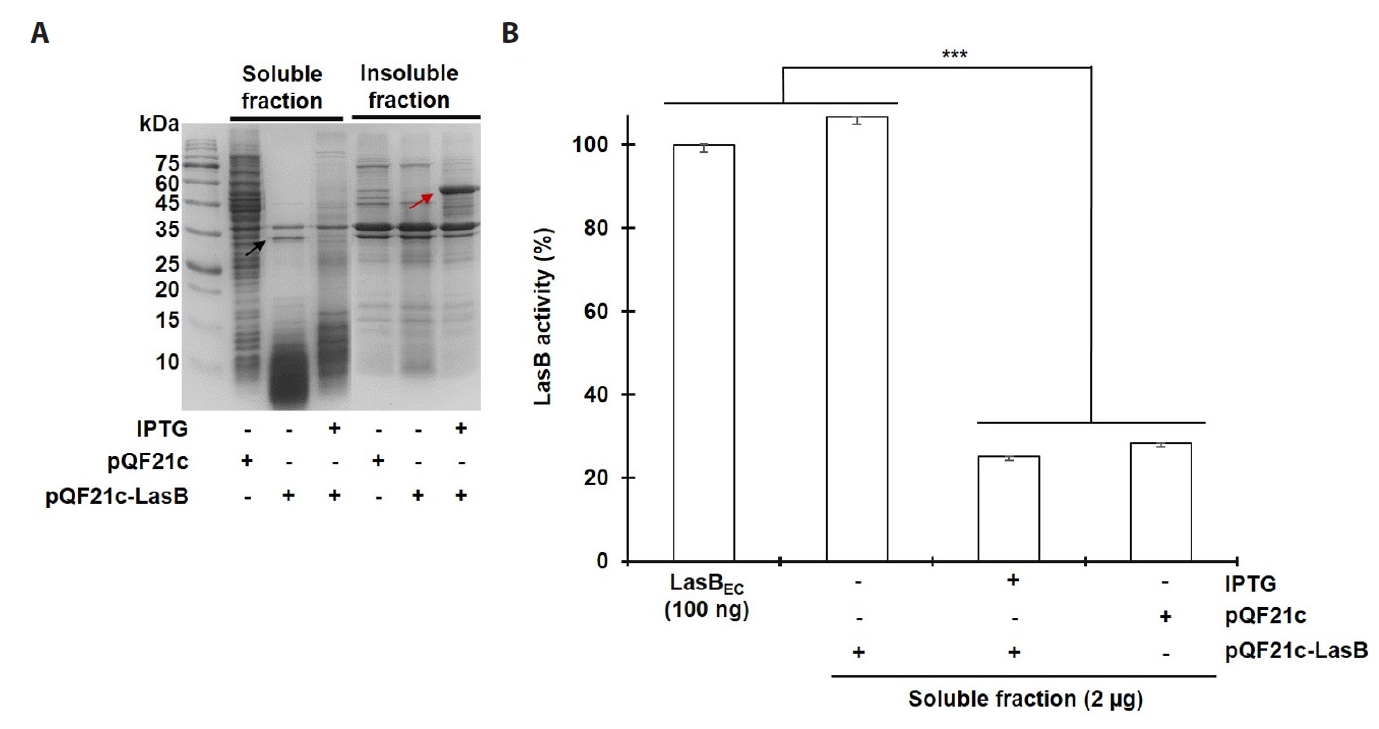
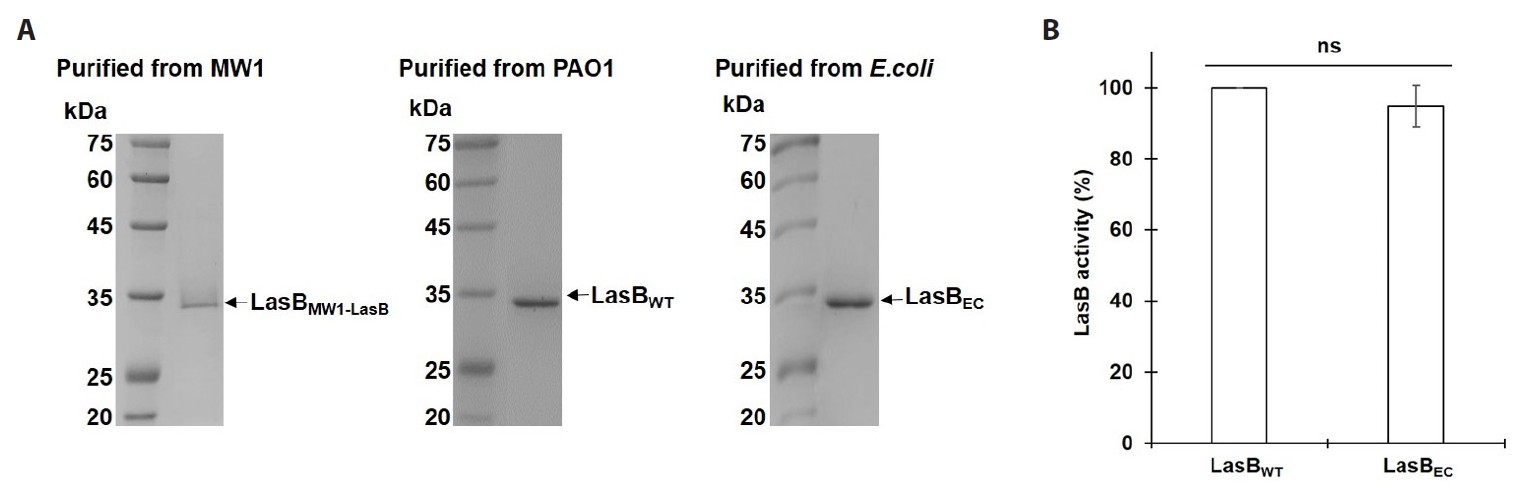

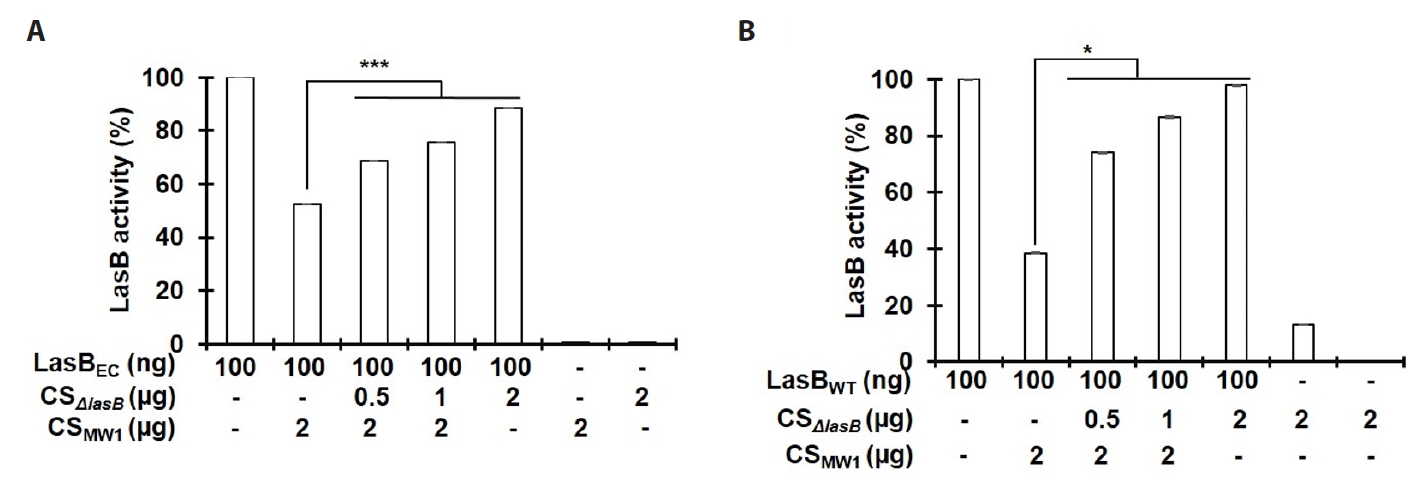
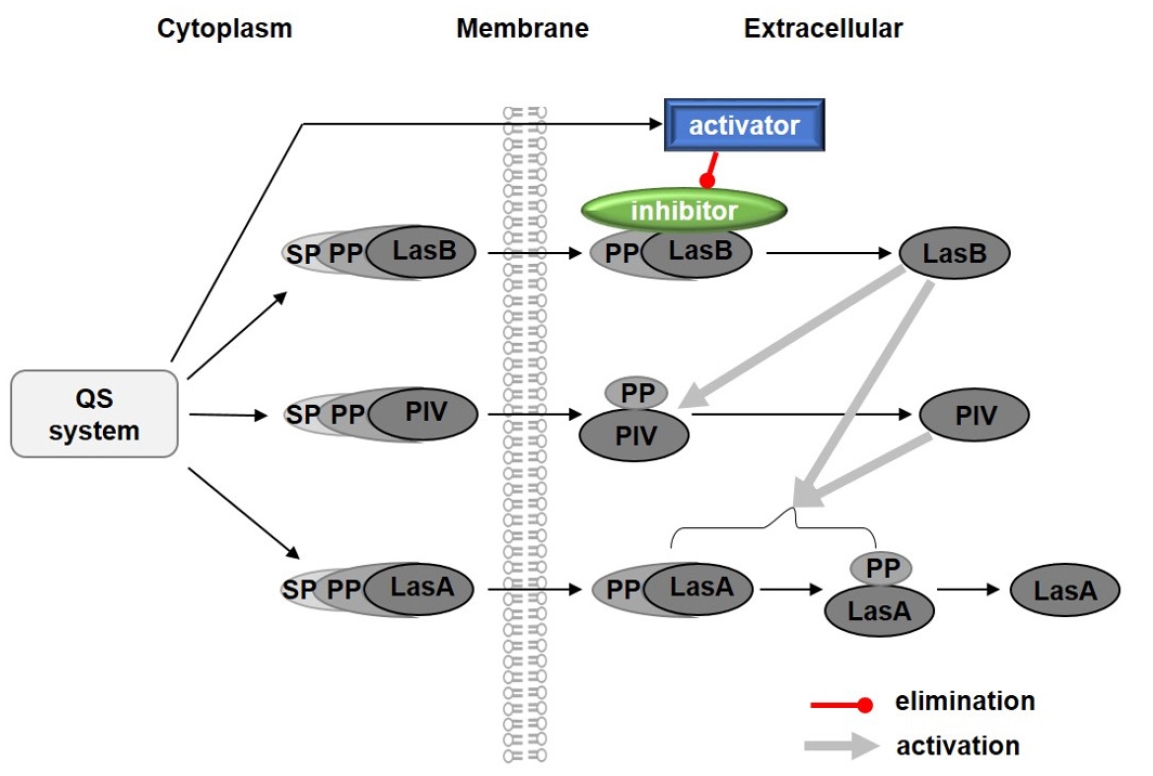
Fig. 1.
Fig. 2.
Fig. 3.
Fig. 4.
Fig. 5.
Fig. 6.
| Name | Description | References |
|---|---|---|
| Pseudomonas aeruginosa | ||
| PAO1 | Wild type P. aeruginosa, QS+ | ( |
| MW1 | lasI-, rhlI- double mutant of PAO1, TcR, QS- | ( |
| ΔlasB | lasB- mutant of PAO1, TcR, QS+ | ( |
| Escherichia coli | ||
| DH5α | supE44ΔlacU169(80lacZΔM15) hsdR17 recA1 gyrA96thi-1 relA1 | Lab. collection |
| BL21(DE3) | F-ompT hsdSB (rB- mB-) dcm gal λ (DE3) | Lab. collection |
| Plasmids | ||
| pJN105 | araC-pBAD promoter fusion plasmid, GmR | ( |
| pSP201 | PA3724 (lasB) in pJN105, GmR | ( |
| pQF21c | Overexpression plasmid for C-terminal histidine-tagged protein (a modified pET21c with a broad-host-range replication origin, Ori1600), replicable in P. aeruginosa, CbR (ApR for E. coli) | ( |
| pQF21c-LasB | lasB in pQF21c, CbR (ApR for E. coli) | ( |
Tc, Tetracycline; Gm, Gentamicin; Cb, Carbenicillin; Ap, Ampicillin; Km, Kanamycin
Table 1.
TOP
 MSK
MSK

 ePub Link
ePub Link Cite this Article
Cite this Article







