Articles
- Page Path
- HOME > J. Microbiol > Volume 63(5); 2025 > Article
-
Full article
Role of the LAMMER kinase LkhA in fungal development and aflatoxin production in Aspergillus flavus - Seong-Hwan Jeong1, He-Jin Cho2, Jae-Hyuk Yu3, Hee-Moon Park4, Hee-Soo Park1,2,3,5,*
-
Journal of Microbiology 2025;63(5):e2503007.
DOI: https://doi.org/10.71150/jm.2503007
Published online: May 27, 2025
1Department of Integrative Biology, Kyungpook National University, Daegu 41566, Republic of Korea
2School of Food Science and Biotechnology, Kyungpook National University, Daegu 41566, Republic of Korea
3Department of Bacteriology, University of Wisconsin-Madison, Madison, Wisconsin 53706, USA
4Department of Microbiology and Molecular Biology, College of Bioscience and Biotechnology, Chungnam National University, Daejeon 34134, Republic of Korea
5Department of Advanced Bioconvergence, Kyungpook National University, Daegu 41566, Republic of Korea
- *Correspondence Hee-Soo Park phsoo97@knu.ac.kr
© The Microbiological Society of Korea
This is an Open Access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/4.0) which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
- 1,030 Views
- 46 Download
ABSTRACT
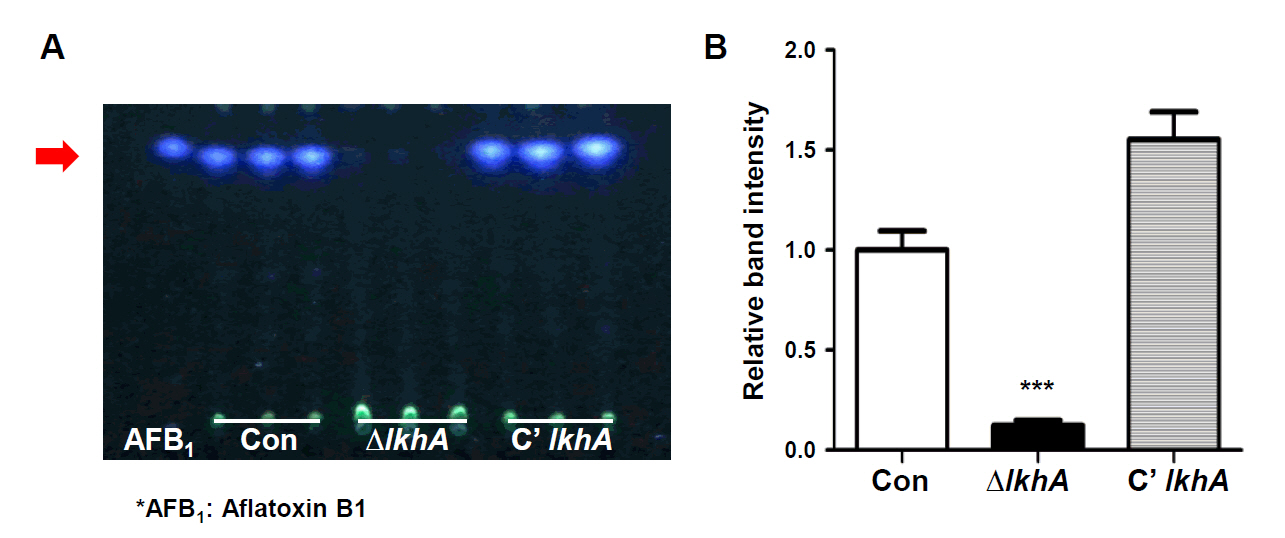
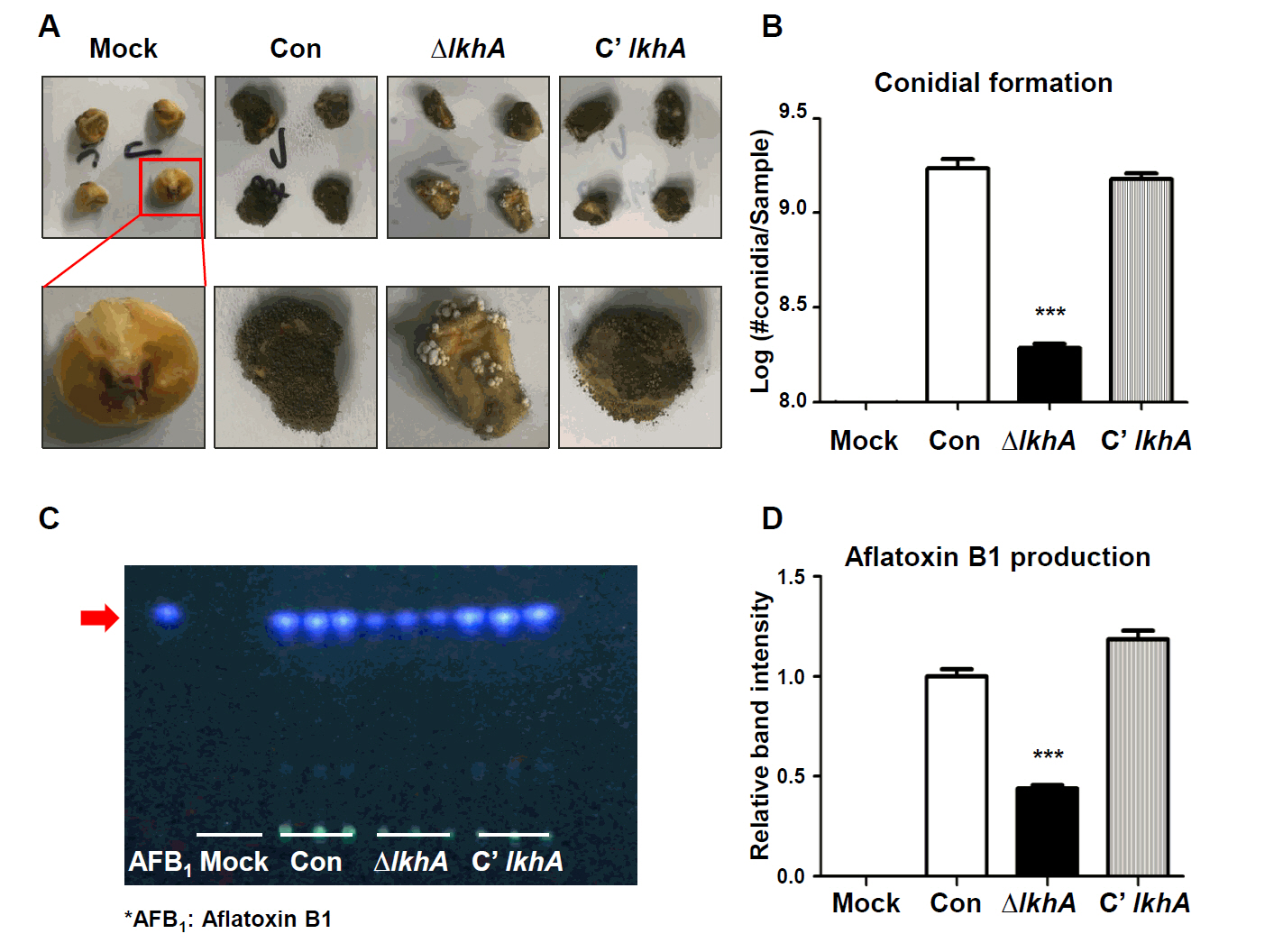
- A well-conserved LAMMER kinase in yeast and filamentous fungi, is a dual-specificity kinase with multiple roles in fungal biology. In this study, we assessed the roles of LkhA in Aspergillus flavus, a toxigenic fungus that produces aflatoxin B1. lkhA deletion mutants exhibited defects in fungal growth, conidiophore development, and sclerotia formation. These mutants exhibited impaired tolerance to oxidative and cell wall stresses. Moreover, the absence of lkhA resulted in a decrease in aflatoxin B1 production. The kernel assay revealed that the lkhA deletion mutants exhibited reduced production of conidia and aflatoxin B1, implying that LkhA can affect fungal toxigenesis and pathogenicity. Taken together, these results demonstrate that LkhA is important for differentiation, mycotoxin production, and pathogenicity in A. flavus.
Introduction
Materials and Methods
Results
Discussion
Acknowledgments
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT) (RS-2024-00339659, 2020R1C1C1004473). This research was supported by the Korea Basic Science Institute (National Research Facilities and Equipment Center) grant funded by the Ministry of Education (2021R1A6C101A416). This research was supported by biological materials Specialized Graduate Program through the Korea Environmental Industry & Technology Institute (KEITI) funded by the Ministry of Environment (MOE).
Conflict of Interest
The authors have no financial conflicts of interest to declare.
| Strain | Relevant genotype | Reference |
|---|---|---|
| NRRL 3357 | A. flavus wild type | ATCC collection |
| NRRL 3357.5 | pyrG− | He et al. (2007) |
| TTJ6.1 | pyrG−; ΔpyrG::AfupyrG+ | Lim et al. (2019) |
| TJSH4.1–3 | pyrG−; ΔlkhA::AfupyrG+ | This study |
| TJSH5.1 and 2 | pyrG−; lkhA(p)::lkhA::FLAG4x::ptrA; ΔlkhA::AfupyrG+ | This study |
| Name | Sequence (5′→3′)a | Purpose |
|---|---|---|
| OHS1542 | CCTGGTCTTTGGTTTGGTACACC | AfupyrG Maker_F |
| OHS1543 | CGACTGGCAGGAGATGATCC | AfupyrG Maker_R |
| OHS2349 | CTACACTTCGATGAGCCTTGC | 5′ AfllkhA DF |
| OHS2350 | GGCTTTGGCCTGTATCATGACTTCA GCAGTGACCGTCGTACTCTAC | 3′ AfllkhA with AfupyrG tail |
| OHS2351 | TTTGGTGACGACAATACCTCCCGAC GTCGTGTTGATCTCAAGTGGC | 5′ AfllkhA with AfupyrG tail |
| OHS2352 | CAGTGAGAACTCGTGCAGG | 3′ AfllkhA DR |
| OHS2353 | CTCTCGGTATTACTGACTACGACG | 5′ AfllkhA NF |
| OHS2354 | GTTCTGCACGACAGTGCAT | 3′ AfllkhA NR |
| OHS2355 | CATCATCAGCAGCACTACGG | 5′ AfllkhA RT_F |
| OHS2356 | CGTAGGGATATGTGGTGGCA | 3′ AfllkhA RT_R |
| OHS2565 | aatt GCGGCCGC CCTACCTCAGATCGACATTGCA | 5′ AfllkhA NotI_F |
| OHS2405 | aatt GCGGCCGC GGTGTTGCGCTGCAACTG | 3′ AfllkhA NotI_R |
| OHS407 | GATATGTCGCCACACTGGAC | AflbrlA RT_F |
| OHS408 | CTGTATTCGCGGCTATTCGG | AflbrlA RT_R |
| OHS409 | CTTCCGCACCTTAACAGCAG | AflabaA RT_F |
| OHS410 | GTTTGCCGGAATTGCCAAAG | AflabaA RT_R |
| OHS405 | TATGTCGGTGATGAGGCACA | Aflactin RT_F |
| OHS406 | AACACGGAGCTCGTTGTAGA | Aflactin RT_R |
- Amaike S, Keller NP. 2011. Aspergillus flavus. Annu Rev Phytopathol. 49: 107–133. ArticlePubMed
- Baltussen TJ, Zoll J, Verweij PE, Melchers WJ. 2020. Molecular mechanisms of conidial germination in Aspergillus spp. Microbiol Mol Biol Rev. 84: e00049–19. ArticlePubMedPDF
- Barratt RW, Johnson GB, Ogata WN. 1965. Wild-type and mutant stocks of Aspergillus nidulans. Genetics. 52: 233–246. ArticlePubMedPMCPDF
- Cho HJ, Son SH, Chen W, Son YE, Lee I, et al. 2022. Regulation of conidiogenesis in Aspergillus flavus. Cells. 11: 2796.ArticlePubMedPMC
- Chowdhury I, Dashi G, Keskitalo S. 2023. Cmgc kinases in health and cancer. Cancers. 15: 3838.ArticlePubMedPMC
- Christensen S, Borrego E, Shim WB, Isakeit T, Kolomiets M. 2012. Quantification of fungal colonization, sporogenesis, and production of mycotoxins using kernel bioassays. J Vis Exp. 62: e3727. Article
- Coley-Smith J, Cooke R. 1971. Survival and germination of fungal sclerotia. Annu Rev Phytopathol. 9: 65–92. Article
- Duan L, Xiao W, Xia F, Liu H, Xiao J, et al. 2016. Two different transcripts of a LAMMER kinase gene play opposite roles in disease resistance. Plant Physiol. 172: 1959–1972. ArticlePubMedPMC
- Dyer PS, O'Gorman CM. 2012. Sexual development and cryptic sexuality in fungi: Insights from Aspergillus species. FEMS Microbiol Rev. 36: 165–192. ArticlePubMed
- Eom TJ, Moon H, Yu JH, Park HS. 2018. Characterization of the velvet regulators in Aspergillus flavus. J Microbiol. 56: 893–901. ArticlePubMedPDF
- Fountain JC, Scully BT, Ni X, Kemerait RC, Lee RD, et al. 2014. Environmental influences on maize-Aspergillus flavus interactions and aflatoxin production. Front Microbiol. 5: 40.ArticlePubMedPMC
- He ZM, Price MS, OBrian GR, Georgianna DR, Payne GA. 2007. Improved protocols for functional analysis in the pathogenic fungus Aspergillus flavus. BMC Microbiol. 7: 104.ArticlePubMedPMCPDF
- Hedayati M, Pasqualotto A, Warn P, Bowyer P, Denning D. 2007. Aspergillus flavus: Human pathogen, allergen and mycotoxin producer. Microbiology. 153: 1677–1692. ArticlePubMed
- Horn BW, Gell RM, Singh R, Sorensen RB, Carbone I. 2016. Sexual reproduction in Aspergillus flavus sclerotia: Acquisition of novel alleles from soil populations and uniparental mitochondrial inheritance. PLoS One. 11: e0146169. ArticlePubMedPMC
- Horn BW, Moore GG, Carbone I. 2009. Sexual reproduction in Aspergillus flavus. Mycologia. 101: 423–429. ArticlePubMed
- Kang EH, Kim Ja, Oh HW, Park HM. 2013. LAMMER kinase LkhA plays multiple roles in the vegetative growth and asexual and sexual development of Aspergillus nidulans. PLoS One. 8: e58762. ArticlePubMedPMC
- Kang WH, Park YD, Hwang JS, Park HM. 2007. RNA-binding protein Csx1 is phosphorylated by LAMMER kinase, Lkh1, in response to oxidative stress in Schizosaccharomyces pombe. FEBS Lett. 581: 3473–3478. ArticlePubMed
- Kang WH, Park YH, Park HM. 2010. The LAMMER kinase homolog, Lkh1, regulates tup transcriptional repressors through phosphorylation in Schizosaccharomyces pombe. J Biol Chem. 285: 13797–13806. ArticlePubMedPMC
- Klich MA. 2007. Aspergillus flavus: The major producer of aflatoxin. Mol Plant Pathol. 8: 713–722. ArticlePubMed
- Krijgsheld P, Bleichrodt Rv, van Veluw G, Wang F, Müller W, et al. 2013. Development in Aspergillus. Stud Mycol. 74: 1–29. ArticlePubMed
- Krishnan S, Manavathu EK, Chandrasekar PH. 2009. Aspergillus flavus: An emerging non-fumigatus Aspergillus species of significance. Mycoses. 52: 206–222. ArticlePubMed
- Kumar A, Pathak H, Bhadauria S, Sudan J. 2021. Aflatoxin contamination in food crops: causes, detection, and management: a review. Food Prod Process Nutr. 3: 17.ArticlePDF
- Kwon S, Choi Y, Kim ES, Lee KT, Bahn YS, et al. 2024. Pleiotropic roles of LAMMER kinase, Lkh1 in stress responses and virulence of Cryptococcus neoformans. Front Cell Infect Microbiol. 14: 1369301.ArticlePubMedPMC
- Lee MK, Kwon NJ, Lee IS, Jung S, Kim SC, et al. 2016. Negative regulation and developmental competence in Aspergillus. Sci Rep. 6: 28874.ArticlePubMedPMCPDF
- Li L, Zhu XM, Wu JQ, Cao N, Bao JD, et al. 2022. The LAMMER kinase MoKns1 regulates growth, conidiation and pathogenicity in Magnaporthe oryzae. Int J Mol Sci. 23: 8104.ArticlePubMedPMC
- Lim JY, Kim YJ, Woo SA, Jeong JW, Lee YR, et al. 2021. The LAMMER kinase, LkhA, affects Aspergillus fumigatus pathogenicity by modulating reproduction and biosynthesis of cell wall PAMPs. Front Cell Infect Microbiol. 11: 756206.ArticlePubMedPMC
- Lim JY, Park HM. 2019. The dual-specificity LAMMER kinase affects stress-response and morphological plasticity in fungi. Front Cell Infect Microbiol. 9: 213.ArticlePubMedPMC
- Lim JY, Park YH, Pyon YH, Yang JM, Yoon JY, et al. 2020. The LAMMER kinase is involved in morphogenesis and response to cell wall- and DNA-damaging stresses in Candida albicans. Med Mycol. 58: 240–247. ArticlePubMedPDF
- Lim SY, Son YE, Lee DH, Eom TJ, Kim MJ, et al. 2019. Function of crzA in fungal development and aflatoxin production in Aspergillus flavus. Toxins (Basel). 11: 567.ArticlePubMedPMC
- Lin L, Wu J, Jiang M, Wang Y. 2021. Plant mitogen-activated protein kinase cascades in environmental stresses. Int J Mol Sci. 22: 1543.ArticlePubMedPMC
- Ojeda-López M, Chen W, Eagle C, Gutiérrez G, Jia W, et al. 2018. Evolution of asexual and sexual reproduction in the aspergilli. Stud Mycol. 91: 37–59. ArticlePubMedPMC
- Park YD, Kang WH, Yang WS, Shin KS, Bae KS, et al. 2003. LAMMER kinase homolog, Lkh1, is involved in oxidative-stress response of fission yeast. Biochem Biophys Res Commun. 311: 1078–1083. ArticlePubMed
- Park HS, Lee MK, Han KH, Kim MJ, Yu JH. 2019. Developmental decisions in Aspergillus nidulans. In Hoffmeister D, Gressler M. (eds.), Biology of the fungal cell. pp. 63–80. Springer, Cham.Article
- Park HS, Ni M, Jeong KC, Kim YH, Yu JH. 2012. The role, interaction and regulation of the Velvet regulator VelB in Aspergillus nidulans. PLoS One. 7: e45935. ArticlePubMedPMC
- Park YH, Park HM. 2011. Temperature sensitivity of sigma background is suppressed by the disruption of ScKNS1 in Saccharomyces cerevisiae. Korean J Microbiol. 47: 167–169.PDF
- Park HS, Yu JH. 2012a. Genetic control of asexual sporulation in filamentous fungi. Curr Opin Microbiol. 15: 669–677. Article
- Park HS, Yu JH. 2012b. Multi-copy genetic screen in Aspergillus nidulans. In Keller N, Turner G. (eds.), Fungal secondary metabolism: methods and protocols, vol. 944, pp. 183–190, Humana Press.Article
- Park HS, Yu JH. 2016. 1 Molecular biology of asexual sporulation in filamentous fungi. In Hoffmeister D. (ed.), The Mycota: Biochemistry and molecular biology, vol. 3, pp. 3–19, Springer, Cham.Article
- Pasqualotto AC. 2009. Differences in pathogenicity and clinical syndromes due to Aspergillus fumigatus and Aspergillus flavus. Med Mycol. 47: S261–S270. ArticlePubMed
- Rushing BR, Selim MI. 2019. Aflatoxin B1: A review on metabolism, toxicity, occurrence in food, occupational exposure, and detoxification methods. Food Chem Toxicol. 124: 81–100. ArticlePubMed
- Savaldi-Goldstein S, Aviv D, Davydov O, Fluhr R. 2003. Alternative splicing modulation by a LAMMER kinase impinges on developmental and transcriptome expression. Plant Cell. 15: 926–938. ArticlePubMedPMC
- Sweany RR, Breunig M, Opoku J, Clay K, Spatafora JW, et al. 2022. Why do plant-pathogenic fungi produce mycotoxins? Potential roles for mycotoxins in the plant ecosystem. Phytopathology. 112: 2044–2051. ArticlePubMed
- Wang F, Sethiya P, Hu X, Guo S, Chen Y, et al. 2021. Transcription in fungal conidia before dormancy produces phenotypically variable conidia that maximize survival in different environments. Nat Microbiol. 6: 1066–1081. ArticlePubMedPDF
- Wicklow D. 1987. Survival of Aspergillus flavus sclerotia in soil. Trans Br Mycol Soc. 83: 131–134. Article
- Yu JH, Hamari Z, Han KH, Seo JA, Reyes-Domínguez Y, et al. 2004. Double-joint PCR: a PCR-based molecular tool for gene manipulations in filamentous fungi. Fungal Genet Biol. 41: 973–981. ArticlePubMed
- Yun B, Farkas R, Lee K, Rabinow L. 1994. The Doa locus encodes a member of a new protein kinase family and is essential for eye and embryonic development in Drosophila melanogaster. Genes Dev. 8: 1160–1173. ArticlePubMed
- Zhao X, Mehrabi R, Xu JR. 2007. Mitogen-activated protein kinase pathways and fungal pathogenesis. Eukaryot Cell. 6: 1701–1714. ArticlePubMedPMCPDF
References
Figure & Data
References
Citations

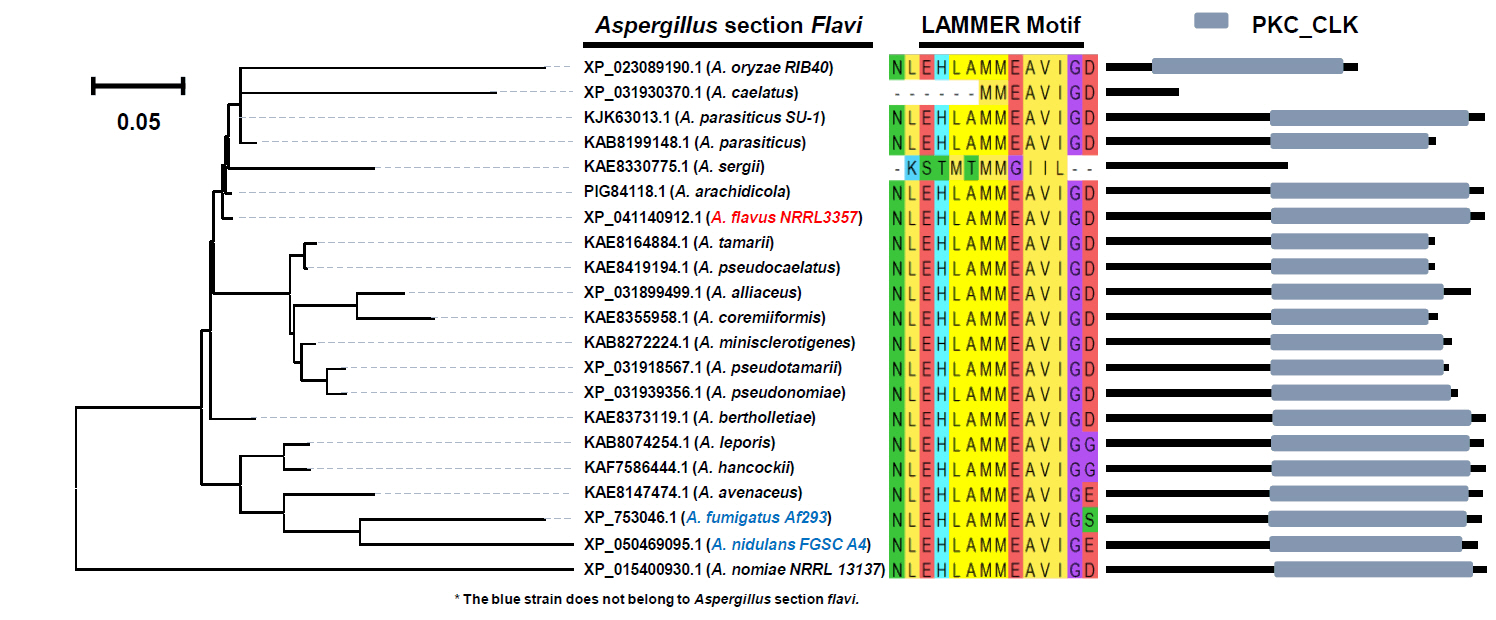
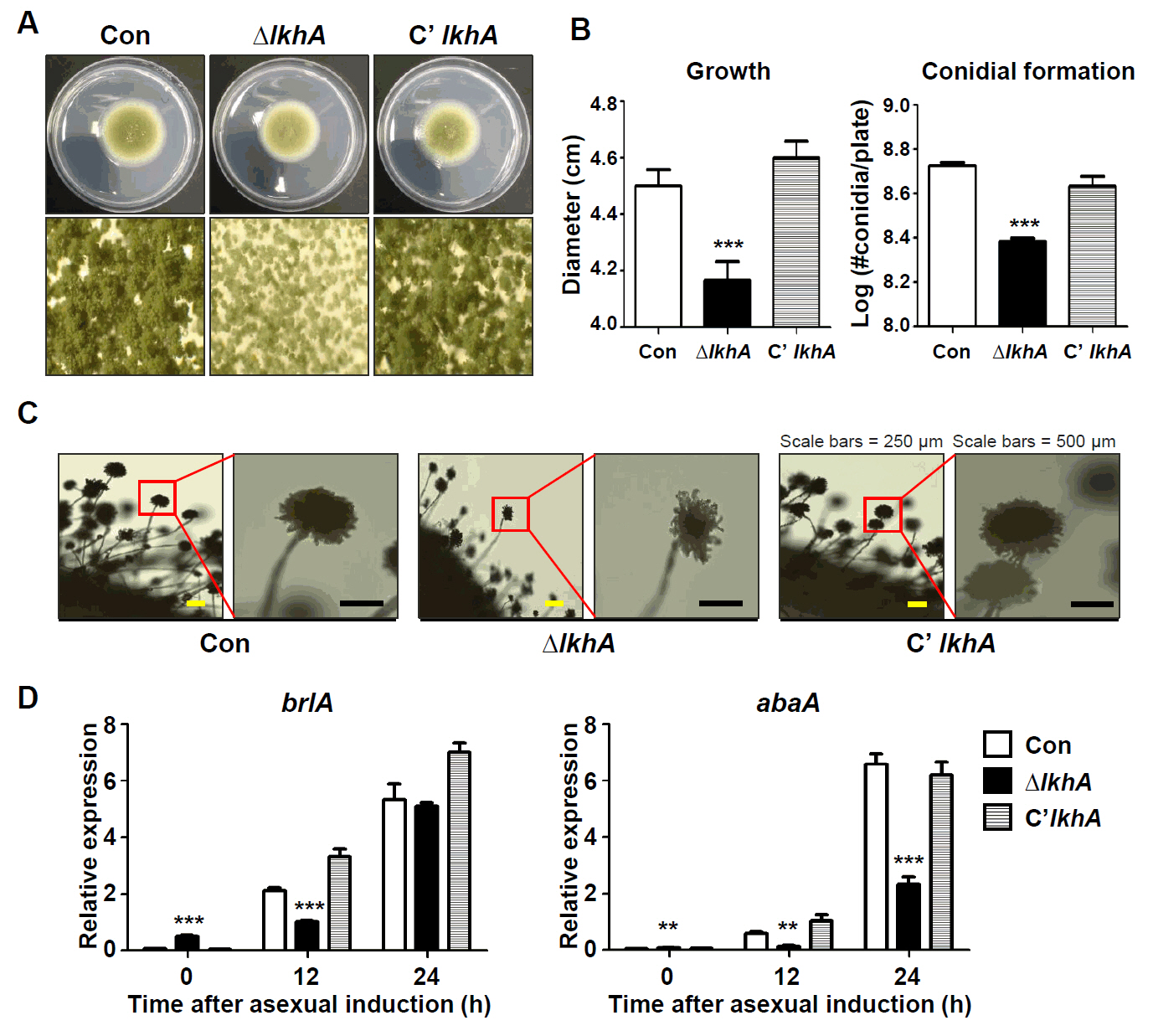
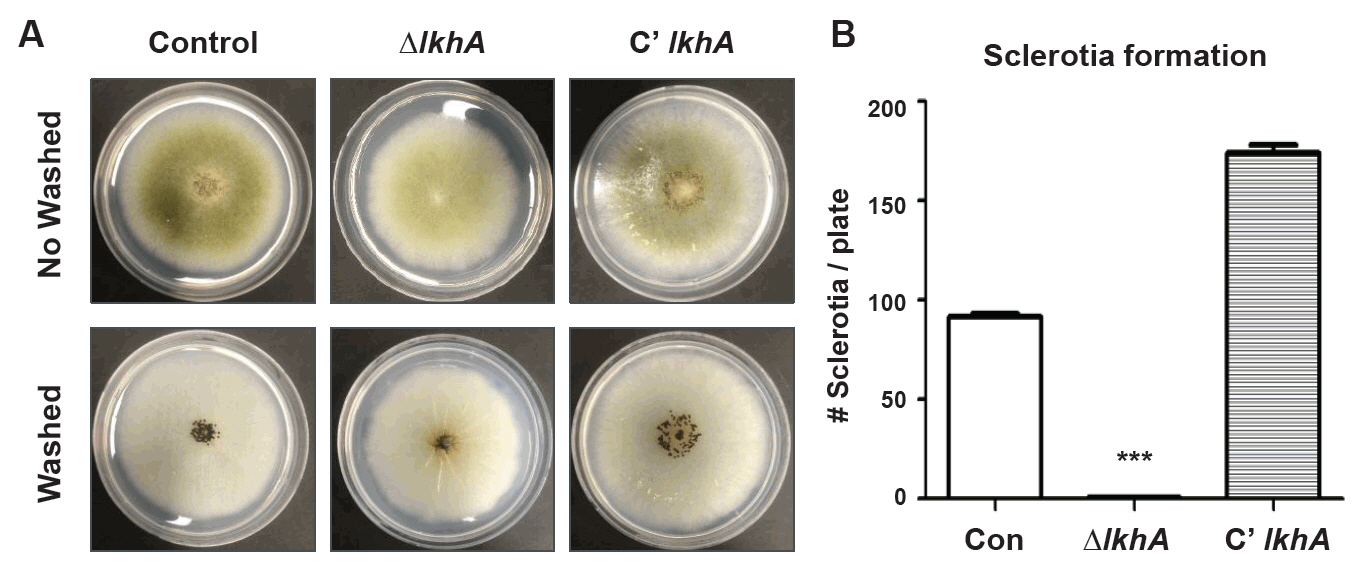
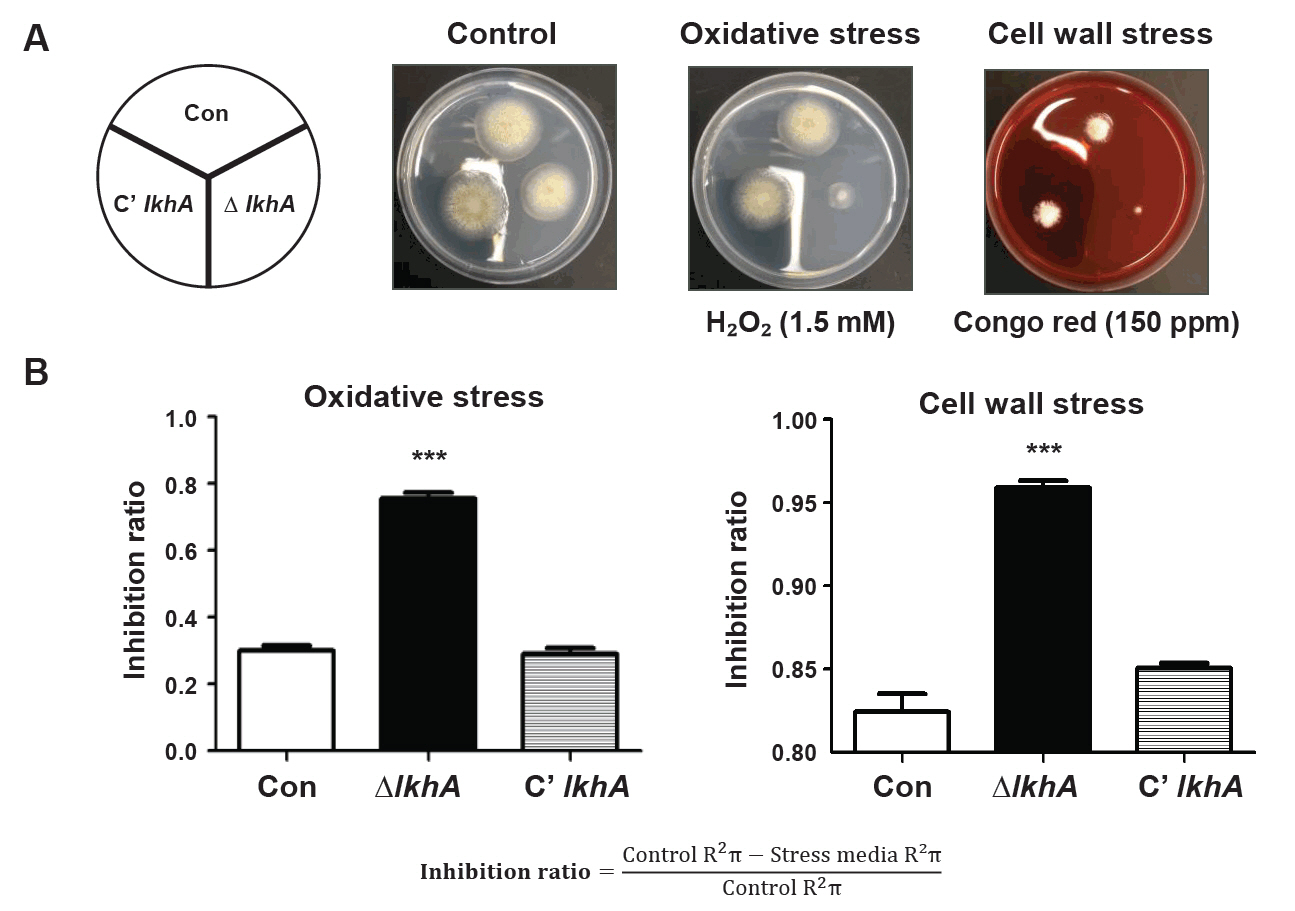
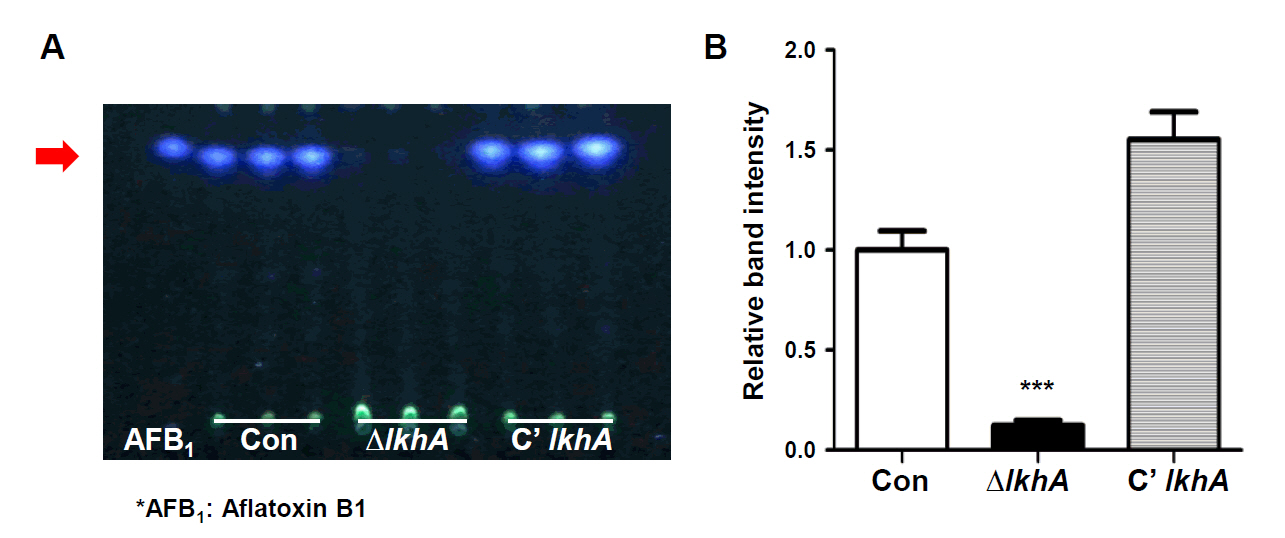
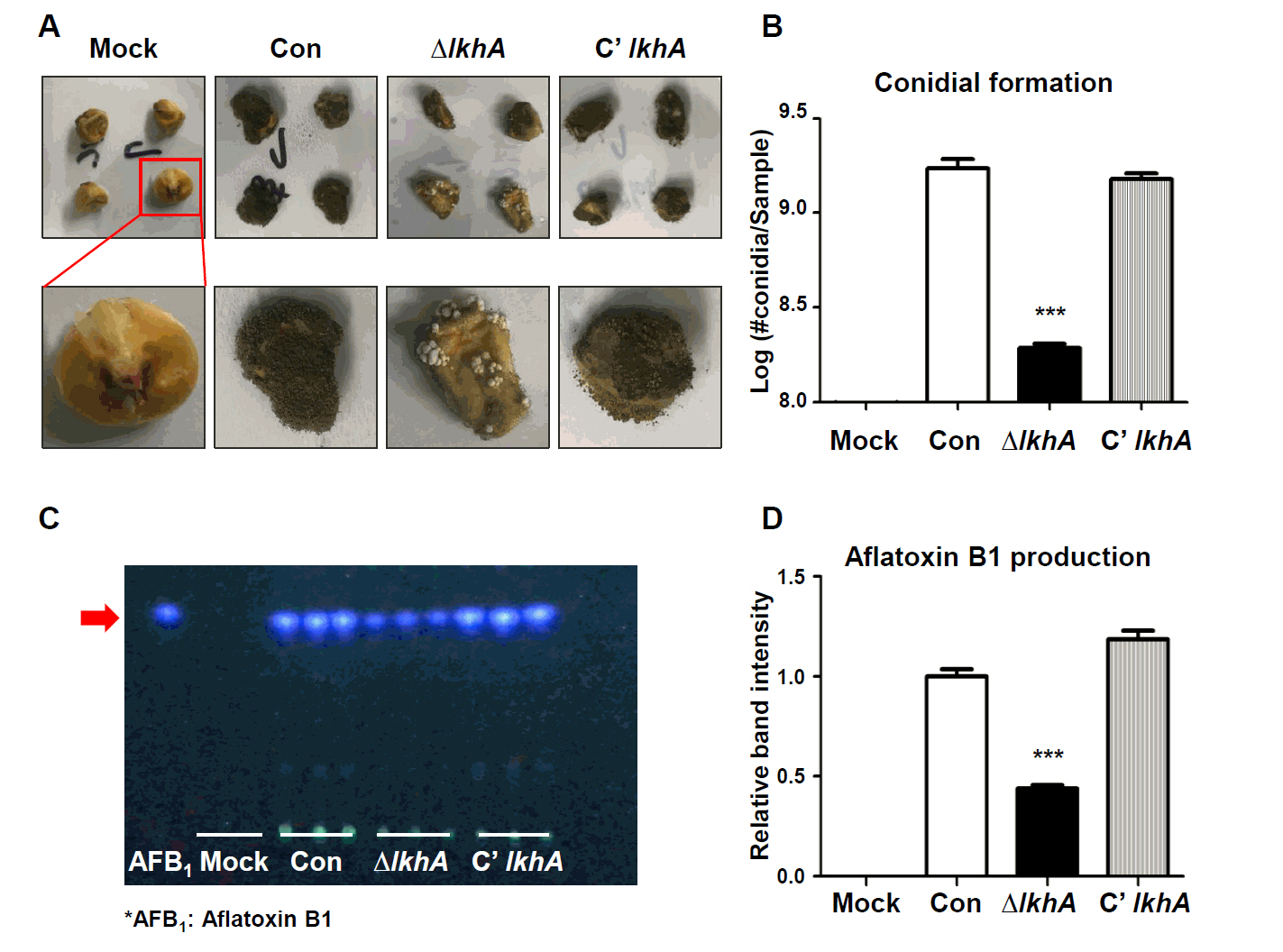
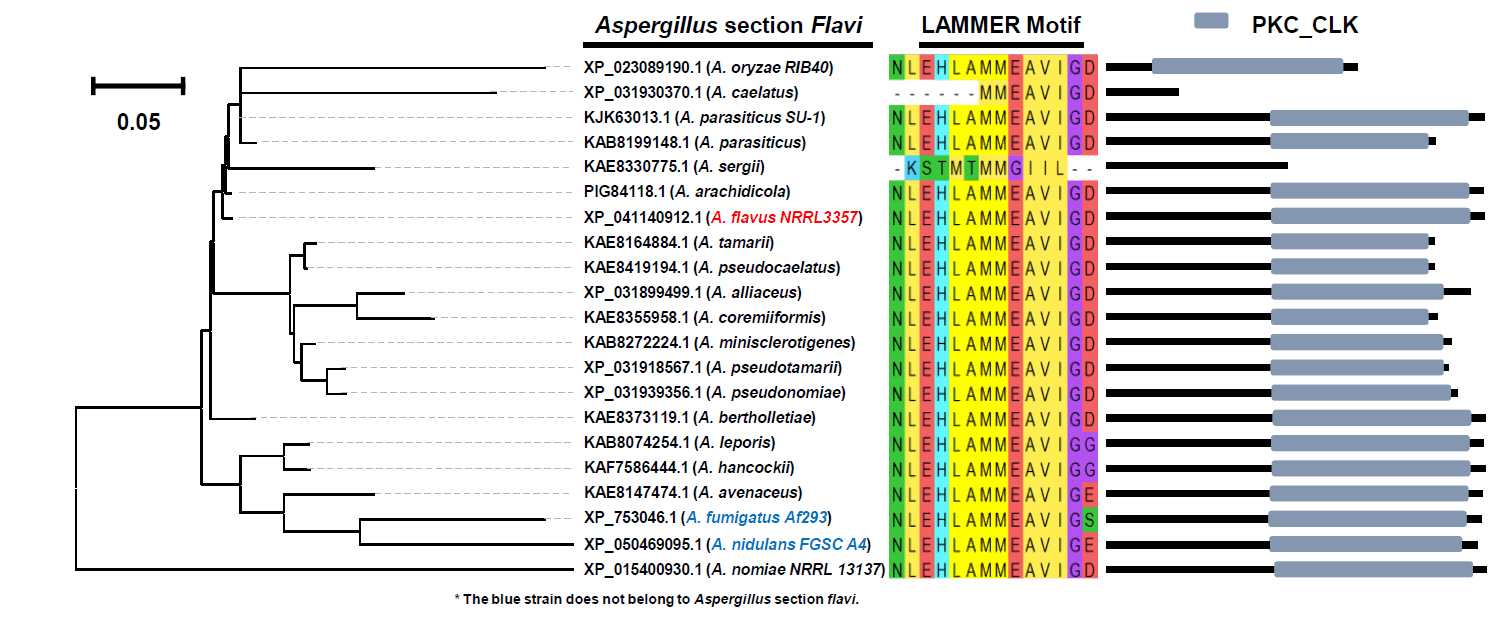
Fig. 1.
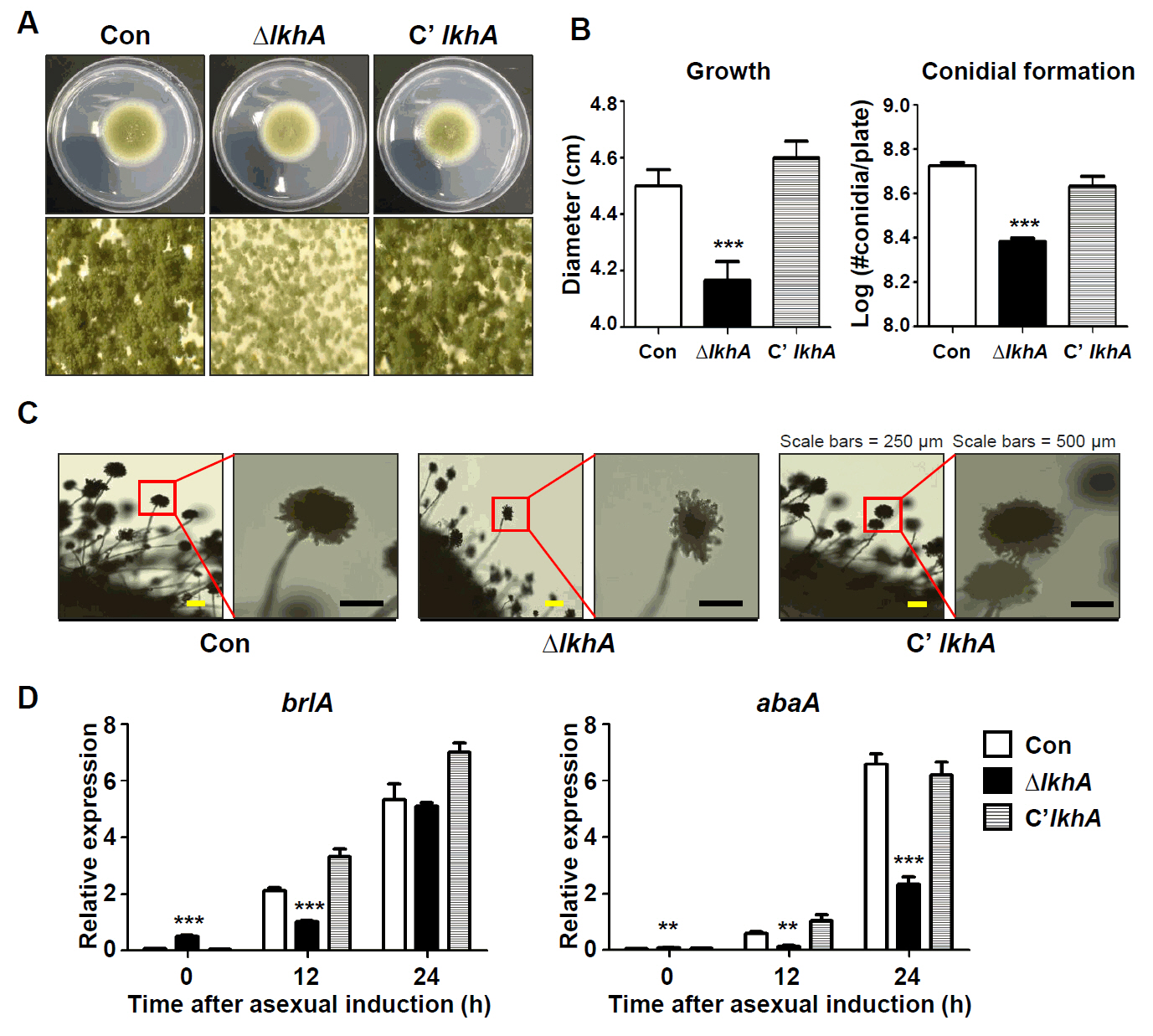
Fig. 2.
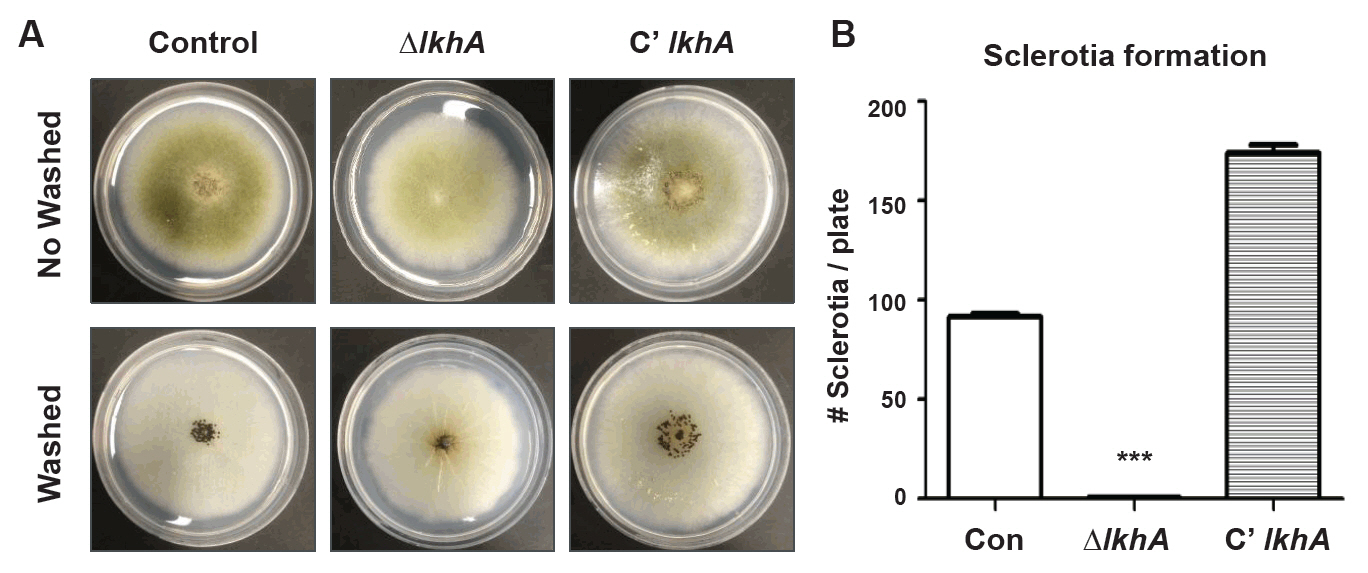
Fig. 3.
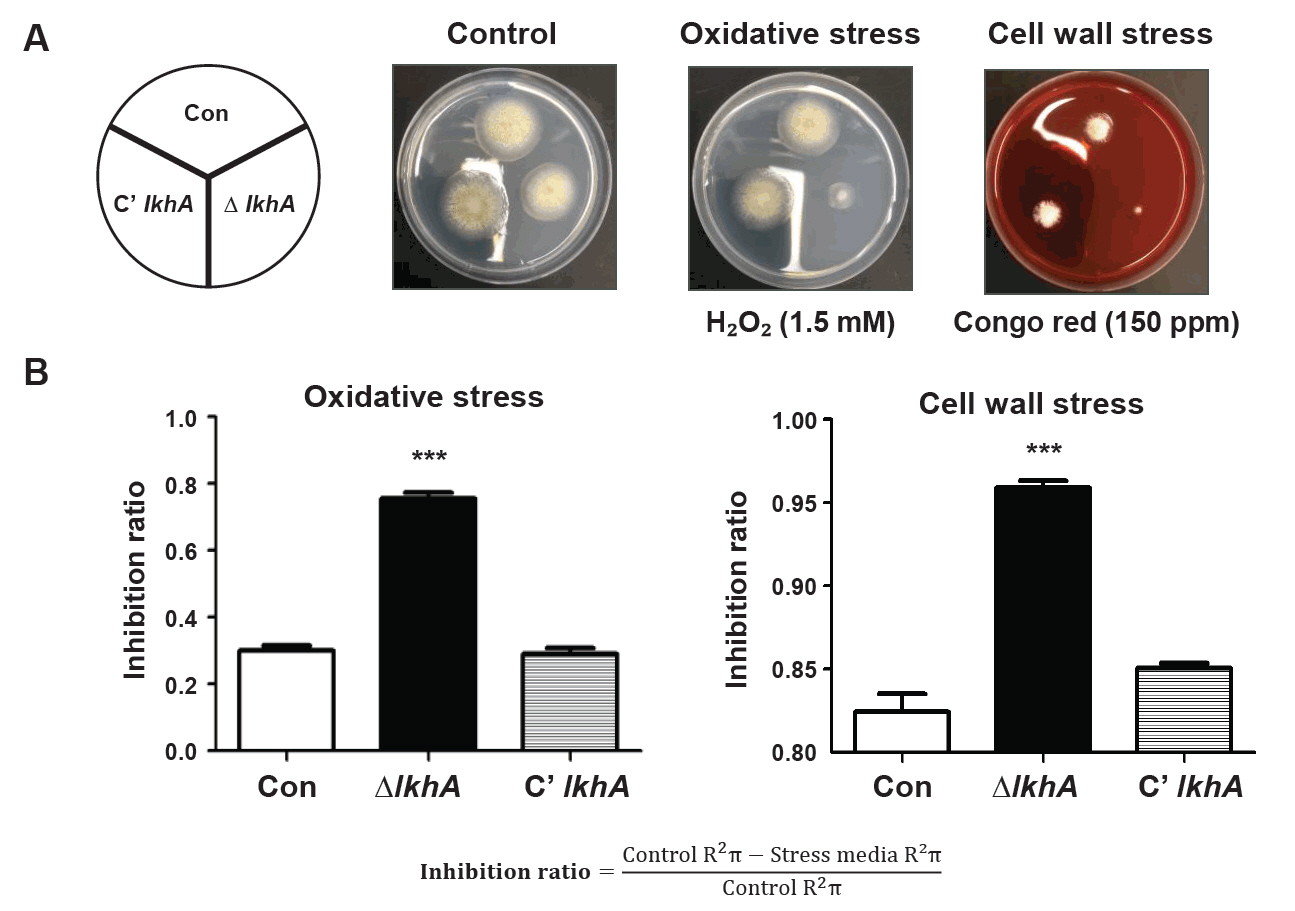
Fig. 4.
Fig. 5.
Fig. 6.
| Strain | Relevant genotype | Reference |
|---|---|---|
| NRRL 3357 | A. flavus wild type | ATCC collection |
| NRRL 3357.5 | pyrG− | |
| TTJ6.1 | pyrG−; ΔpyrG::AfupyrG+ | |
| TJSH4.1–3 | pyrG−; ΔlkhA::AfupyrG+ | This study |
| TJSH5.1 and 2 | pyrG−; lkhA(p)::lkhA::FLAG4x::ptrA; ΔlkhA::AfupyrG+ | This study |
| Name | Sequence (5′→3′) |
Purpose |
|---|---|---|
| OHS1542 | CCTGGTCTTTGGTTTGGTACACC | AfupyrG Maker_F |
| OHS1543 | CGACTGGCAGGAGATGATCC | AfupyrG Maker_R |
| OHS2349 | CTACACTTCGATGAGCCTTGC | 5′ AfllkhA DF |
| OHS2350 | GGCTTTGGCCTGTATCATGACTTCA GCAGTGACCGTCGTACTCTAC | 3′ AfllkhA with AfupyrG tail |
| OHS2351 | TTTGGTGACGACAATACCTCCCGAC GTCGTGTTGATCTCAAGTGGC | 5′ AfllkhA with AfupyrG tail |
| OHS2352 | CAGTGAGAACTCGTGCAGG | 3′ AfllkhA DR |
| OHS2353 | CTCTCGGTATTACTGACTACGACG | 5′ AfllkhA NF |
| OHS2354 | GTTCTGCACGACAGTGCAT | 3′ AfllkhA NR |
| OHS2355 | CATCATCAGCAGCACTACGG | 5′ AfllkhA RT_F |
| OHS2356 | CGTAGGGATATGTGGTGGCA | 3′ AfllkhA RT_R |
| OHS2565 | aatt GCGGCCGC CCTACCTCAGATCGACATTGCA | 5′ AfllkhA NotI_F |
| OHS2405 | aatt GCGGCCGC GGTGTTGCGCTGCAACTG | 3′ AfllkhA NotI_R |
| OHS407 | GATATGTCGCCACACTGGAC | AflbrlA RT_F |
| OHS408 | CTGTATTCGCGGCTATTCGG | AflbrlA RT_R |
| OHS409 | CTTCCGCACCTTAACAGCAG | AflabaA RT_F |
| OHS410 | GTTTGCCGGAATTGCCAAAG | AflabaA RT_R |
| OHS405 | TATGTCGGTGATGAGGCACA | Aflactin RT_F |
| OHS406 | AACACGGAGCTCGTTGTAGA | Aflactin RT_R |
Tail sequences are shown in italics. Restriction enzyme sites are presented in bold.
Table 1.
Table 2.
TOP
 MSK
MSK

 ePub Link
ePub Link Cite this Article
Cite this Article







