Articles
- Page Path
- HOME > J. Microbiol > Volume 63(2); 2025 > Article
-
Full article
Functional importance of Ser323 in cysteine desulfhydrase and cystathionine gamma-lyase MccB of Staphylococcus aureus -
Dukwon Lee1, Hyojeong Lee1, Kyumi Byun2, Eun-Su Park3, Nam-Chul Ha1,4,*

-
Journal of Microbiology 2025;63(2):e2411026.
DOI: https://doi.org/10.71150/jm.2411026
Published online: February 27, 2025
1Research Institute of Agriculture and Life Sciences, Department of Agricultural Biotechnology, CALS, Seoul National University, Seoul 08826, Republic of Korea
2School of Biological Sciences, Institute of Molecular Biology and Genetics, Seoul National University, Seoul 08826, Republic of Korea
3The Internship Program at the Food Biochemistry Laboratory, Department of Agricultural Biotechnology, CALS, Seoul National University, Seoul 08826, Republic of Korea
4Center for Food and Bioconvergence, Interdisciplinary Programs in Agricultural Genomics, CALS, Seoul National University, Seoul 08826, Republic of Korea
- *Correspondence Nam-Chul Ha hanc210@snu.ac.kr
© The Microbiological Society of Korea
This is an Open Access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/4.0) which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
ABSTRACT
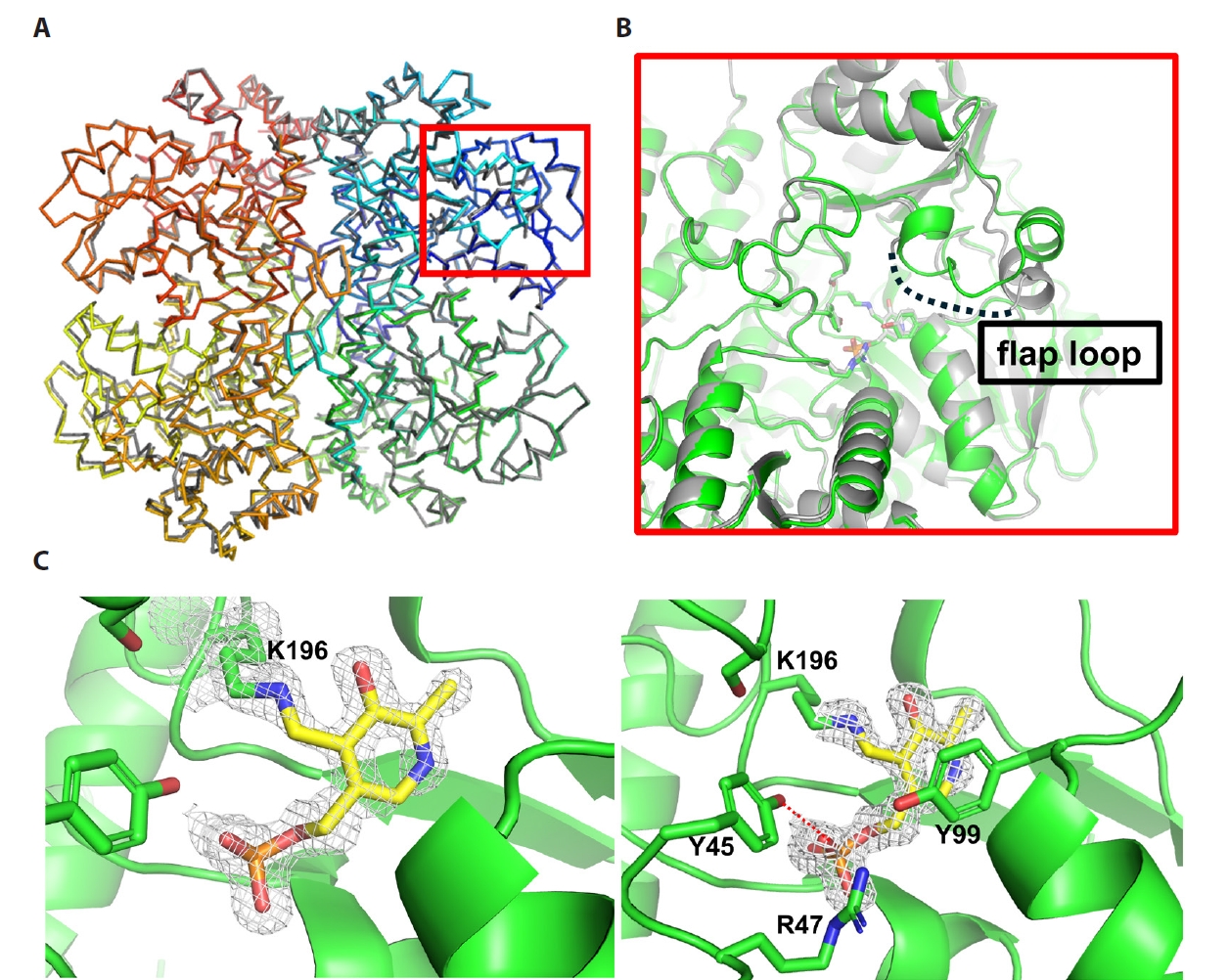
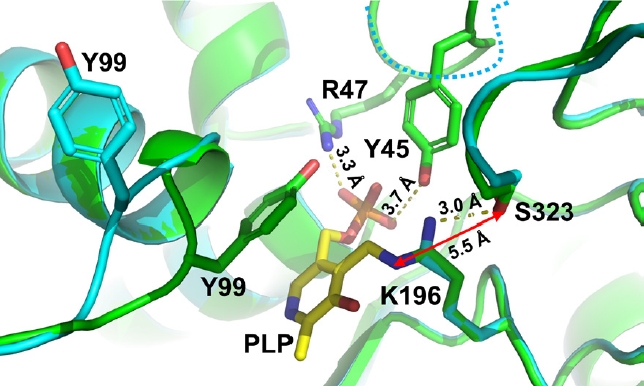
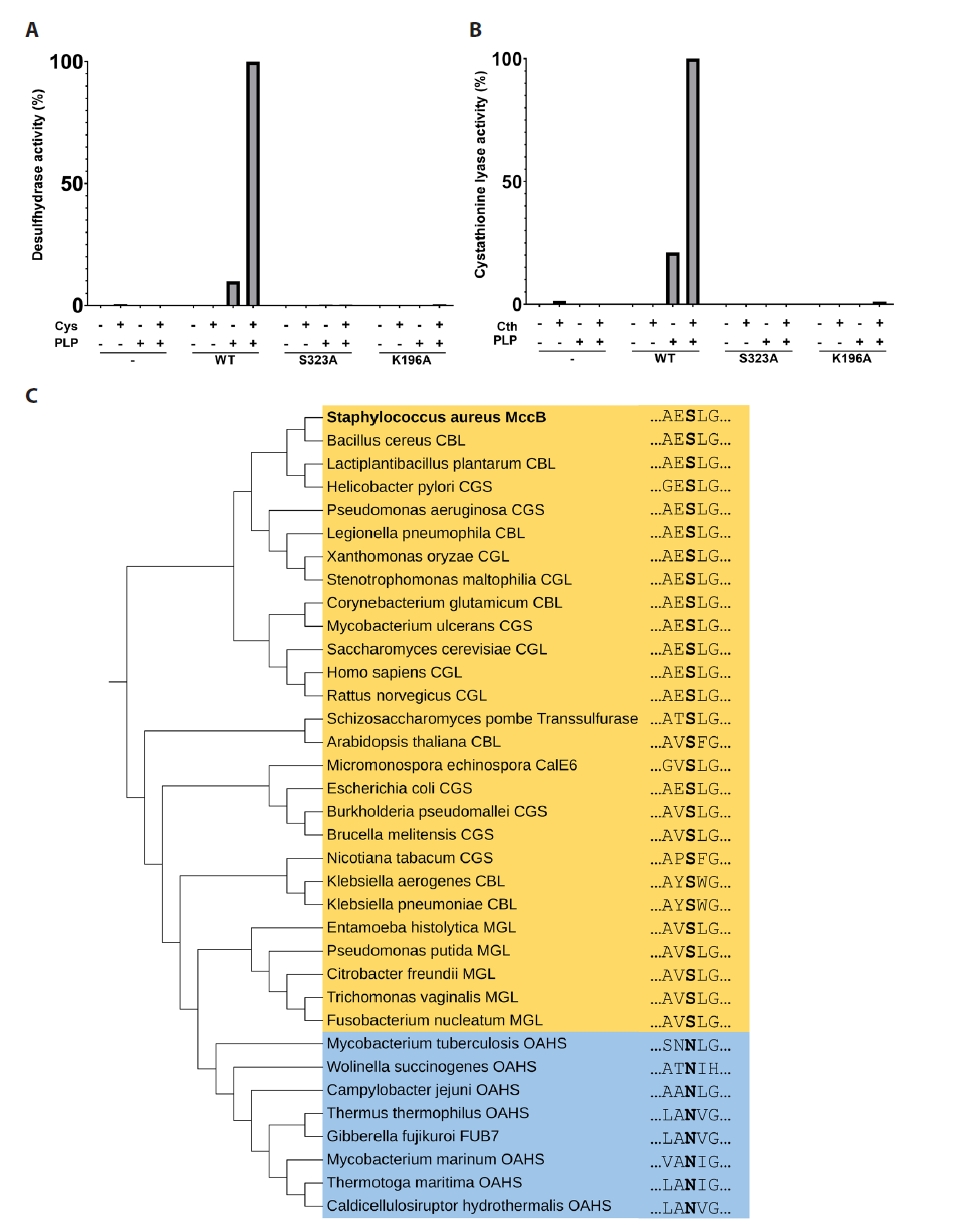
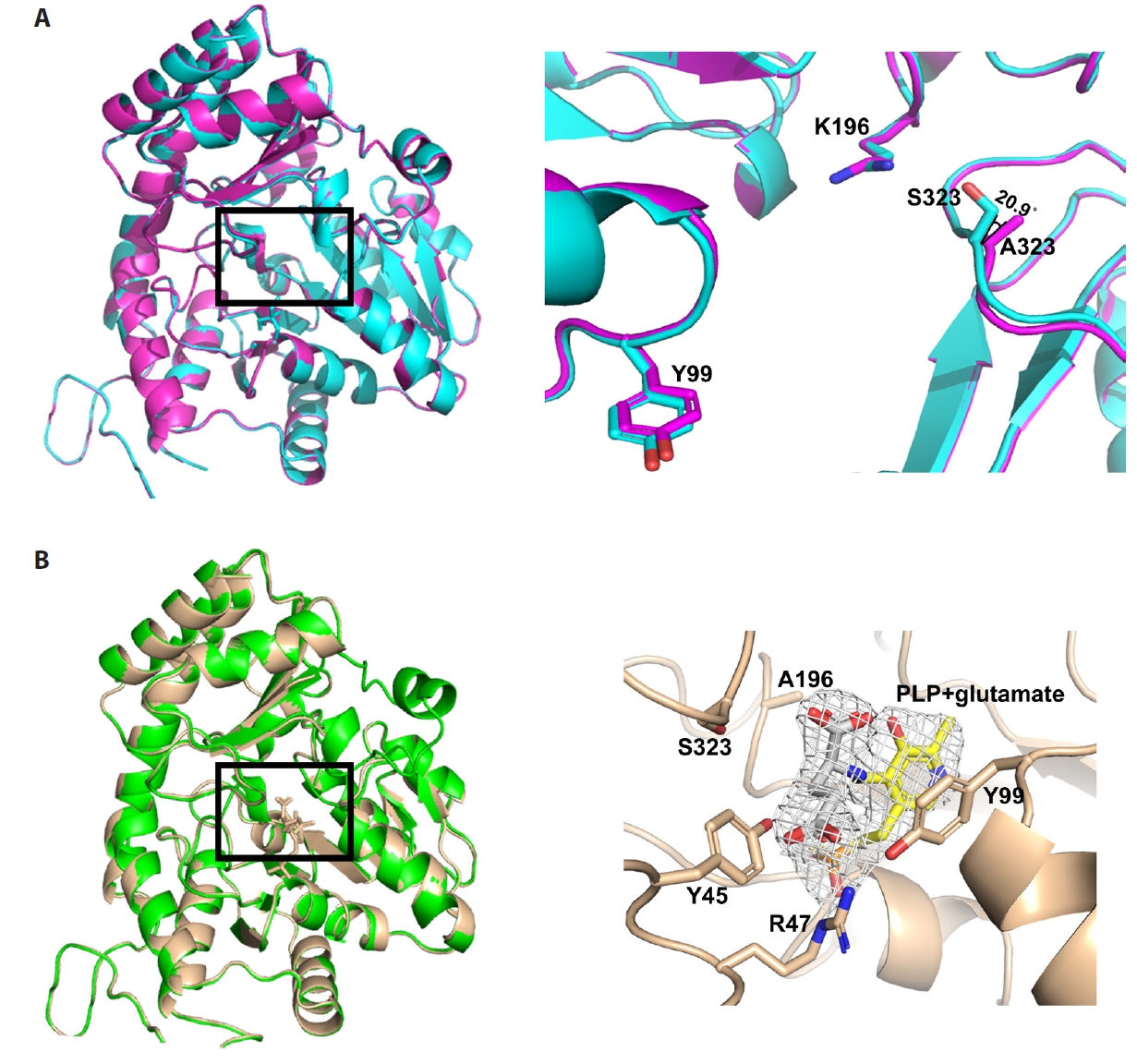
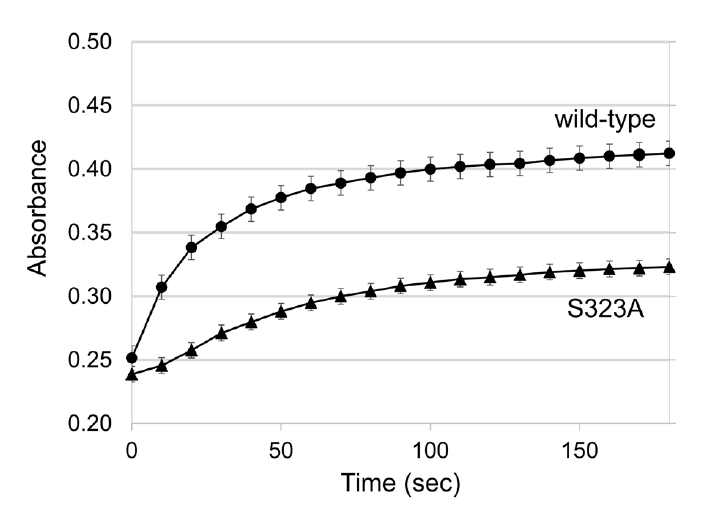
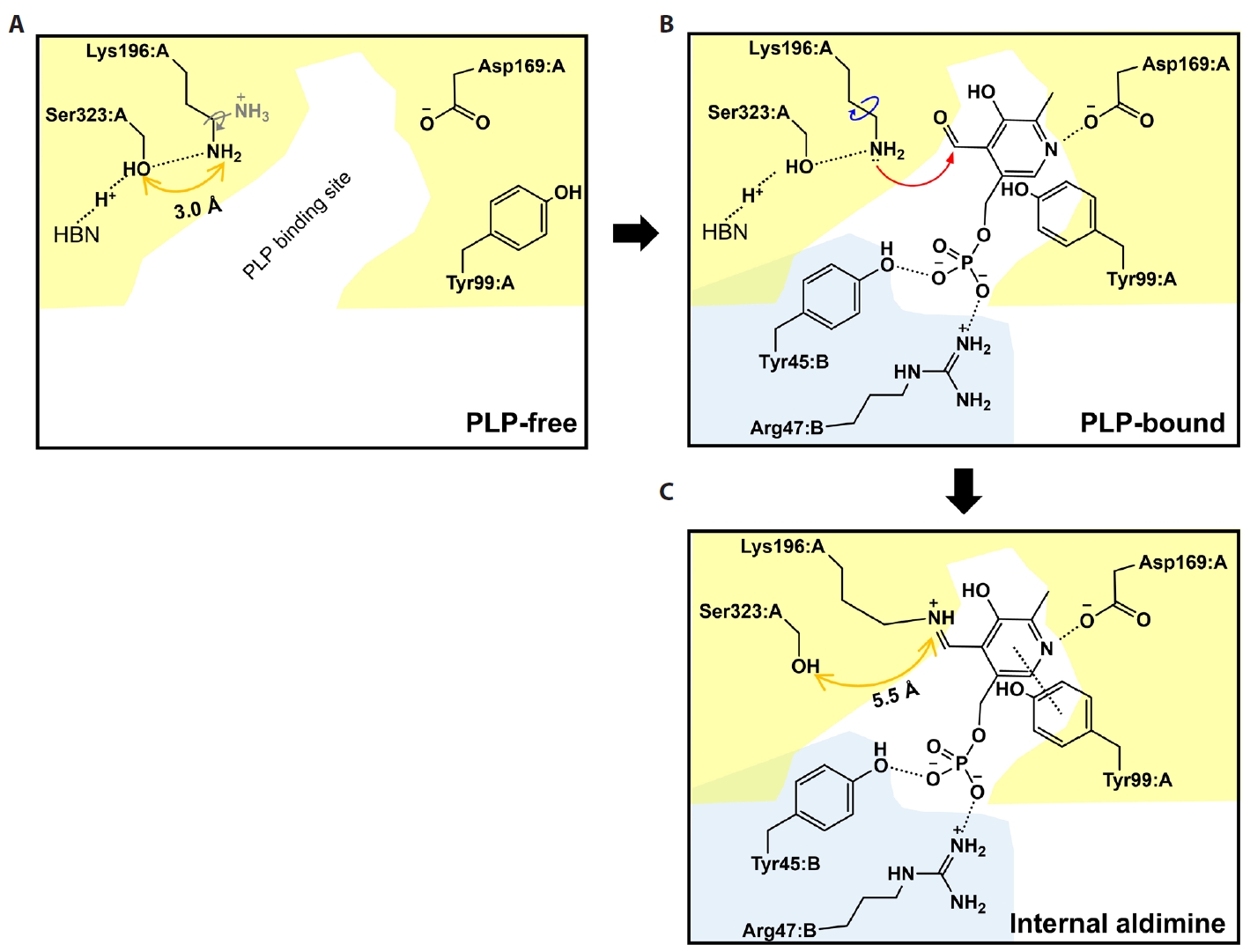
- Pyridoxal 5'-phosphate (PLP)-dependent enzymes participate in various reactions involved in methionine and cysteine metabolism. The representative foodborne pathogen Staphylococcus aureus expresses the PLP-dependent enzyme MccB, which exhibits both cystathionine gamma-lyase (CGL) and cysteine desulfhydrase activities. In this study, we investigated the role of Ser323 in MccB, a conserved residue in many PLP-dependent enzymes in the transsulfuration pathway. Our findings reveal that Ser323 forms a hydrogen bond with the catalytic lysine in the absence of PLP, and upon internal aldimine formation, PLP-bound lysine is repositioned away from Ser323. Substituting Ser323 with alanine abolishes the enzymatic activity, similar to mutations at the catalytic lysine site. Spectroscopic analysis suggests that Ser323 is essential for the rapid formation of the internal aldimine with lysine in wild-type MccB. This study highlights the crucial role of Ser323 in catalysis, with broader implications for other PLP-dependent enzymes, and enhances our understanding of the molecular mechanisms involved in the selective control of foodborne pathogenic bacteria.
Introduction
Materials and Methods
Results
Discussion
Acknowledgments
This research was supported by the Korea Institute of Planning and Evaluation for Technology in Food, Agriculture, Forestry, and Fisheries (IPET), funded by the Ministry of Agriculture, Food, and Rural Affairs (MAFRA) (RS-2021-IP321036 and RS-2024-00402136 to N.C.H.). This research was supported by the Bio & Medical Technology Development Program of the National Research Foundation (NRF), funded by the Ministry of Science & ICT (MSIT) (2022R1A2C1091783 and 2019M3E5D606387122 to N.C.H.). This research was also supported by the Bio & Medical Technology Development Program of the NRF funded by the MSIT (RS-2024-00344154 to N.C.H.). We made use of Beamline 5C at Pohang Accelerator Laboratory (Pohang, Republic of Korea)
Conflict of interest
The authors declare that they have no conflicts of interest regarding the content of this article.
Ethical Statements
This study did not involve human participants, animal subjects, or biological materials requiring specific ethical approval. The research was conducted using bacterial proteins expressed in a heterologous bacterial host system. All experiments were carried out in accordance with institutional and national guidelines for biosafety and the responsible use of microorganisms.
Supplementary Information
Fig. S1.
Fig. S2.
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, et al. 2010. Phenix: A comprehensive python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 66(2): 213–221. ArticlePubMedPMC
- Binkley F, Okeson D. 1950. Purification of the enzyme responsible for the cleavage of cystathionine. J Biol Chem. 182(1): 273–277. Article
- Byun K, Lee D, Kim H, Lee DH, Xu Y, et al. 2024. EGCG inhibits cystathionine gamma-lyase MccB from Staphylococcus aureus by making a hemiacetal compound with pyridoxal phosphate. Food Biosci. 57: 103560.Article
- Du YL, Ryan KS. 2019. Pyridoxal phosphate-dependent reactions in the biosynthesis of natural products. Nat Prod Rep. 36(3): 430–457. ArticlePubMed
- Emsley P, Cowtan K. 2004. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 60(12): 2126–2132. ArticlePubMed
- Evande R, Ojha S, Banerjee R. 2004. Visualization of PLP-bound intermediates in hemeless variants of human cystathionine β-synthase: Evidence that lysine 119 is a general base. Arch Biochem Biophys. 427(2): 188–196. ArticlePubMed
- Gophna U, Bapteste E, Doolittle WF, Biran D, and Ron EZ. 2005. Evolutionary plasticity of methionine biosynthesis. Gene. 355: 48–57. ArticlePubMed
- Grishin NV, Phillips MA, Goldsmith EJ. 1995. Modeling of the spatial structure of eukaryotic ornithine decarboxylases. Protein Sci. 4(7): 1291–1304. ArticlePubMedPMC
- Han L, Schwabacher AW, Moran GR, Silvaggi NR. 2015. Streptomyces wadayamensis MppP is a pyridoxal 5′-phosphate-dependent l-arginine α-deaminase, γ-hydroxylase in the enduracididine biosynthetic pathway. Biochemistry. 54(47): 7029–7040. ArticlePubMed
- Hayashi H. 1995. Pyridoxal enzymes: Mechanistic diversity and uniformity. J Biochem. 118(3): 463–473. ArticlePubMed
- Huai Q, Xia Y, Chen Y, Callahan B, Li N, et al. 2001. Crystal structures of 1-aminocyclopropane-1-carboxylate (ACC) synthase in complex with aminoethoxyvinylglycine and pyridoxal-5′-phosphate provide new insight into catalytic mechanisms. J Biol Chem. 276(41): 38210–38216. ArticlePubMed
- Hullo MF, Auger S, Soutourina O, Barzu O, Yvon M, et al. 2007. Conversion of methionine to cysteine in Bacillus subtilis and its regulation. J Bacteriol. 189(1): 187–197. ArticlePubMedPDF
- Jeong S. 2023. Function and regulation of nitric oxide signaling in Drosophila. Mol Cells. 47(1): 100006.ArticlePubMedPMC
- Kraus JP, Hašek J, Kožich V, Collard R, Venezia S, et al. 2009. Cystathionine γ-lyase: Clinical, metabolic, genetic, and structural studies. Mol Genet Metab. 97(4): 250–259. ArticlePubMedPMC
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6): 1547–1549. ArticlePubMedPMC
- Le Loir Y, Baron F, Gautier M. 2003. Staphylococcus aureus and food poisoning. Genet Mol Res. 2(1): 63–76. PubMed
- Lee D, Jeong S, Ahn J, Ha NC, Kwon AR. 2019. Crystal structure of bacterial cystathionine γ-lyase in the cysteine biosynthesis pathway of Staphylococcus aureus. Crystals. 9(12): 656.Article
- Lowy FD. 1998. Staphylococcus aureus infections. N Engl J Med. 339(8): 520–532. ArticlePubMed
- Luhachack L, Nudler E. 2014. Bacterial gasotransmitters: An innate defense against antibiotics. Curr Opin Microbiol. 21: 13–17. ArticlePubMed
- Morino Y, Nagashima F. 1984. Pyridoxal phosphate-binding site in enzymes: Reduction and comparison of sequences, pp. 116–137. Methods in enzymology, Elsevier.PDF
- Otwinowski Z, Minor W. 1997. Processing of X-ray diffraction data collected in oscillation mode, pp. 307–326. Methods in enzymology, Elsevier.PDF
- Park SY, Ha SC, Kim YG. 2017. The protein crystallography beamlines at the pohang light source II. Biodesign. 5(1): 30–34.PDF
- Schneider G, Käck H, Lindqvist Y. 2000. The manifold of vitamin B6 dependent enzymes. Structure. 8(1): R1–R6. ArticlePubMed
- Shatalin K, Nuthanakanti A, Kaushik A, Shishov D, Peselis A, et al. 2021. Inhibitors of bacterial H2S biogenesis targeting antibiotic resistance and tolerance. Science. 372(6547): 1169–1175. ArticlePubMedPMC
- Song HK. 2023. Quo vadis experimental structural biology? Mol Cells. 46(2): 71–73. ArticlePubMedPMC
- Toney MD. 2011. Controlling reaction specificity in pyridoxal phosphate enzymes. Biochim Biophys Acta. 1814(11): 1407–1418. ArticlePubMedPMC
- Tramonti A, Ghatge MS, Babor JT, Musayev FN, di Salvo ML, et al. 2022. Characterization of the Escherichia coli pyridoxal 5′‐phosphate homeostasis protein (YggS): Role of lysine residues in PLP binding and protein stability. Protein Sci. 31(11): e4471. ArticlePubMedPMC
- Vacca RA, Giannattasio S, Capitani G, Marra E, Christen P. 2008. Molecular evolution of B6 enzymes: Binding of pyridoxal-5'-phosphate and Lys41Arg substitution turn ribonuclease a into A model B6 protoenzyme. BMC Biochem. 9: 17.ArticlePubMedPMCPDF
- Wang R. 2012. Physiological implications of hydrogen sulfide: A whiff exploration that blossomed. Physiol Rev. 92(2): 791–896. ArticlePubMed
- Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, et al. 2011. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 67(Pt 4): 235–242. ArticlePubMedPMC
References
Figure & Data
References
Citations

- Switch of cupreous valance within copper-contained silicocarnotite enhancing anti-bacterial and osteogenic properties in treating infected bone defects
Hongyu Chen, Xin Wang, Zizhuo Liu, Yi Wang, Lei Shi, Weida Li, Xiude Chen, Ye Sun, Shunxiang Xu, Zuyan Lu, Yaokai Gan, Fanyan Deng, Qiang Wu
Chemical Engineering Journal.2026; 527: 172134. CrossRef
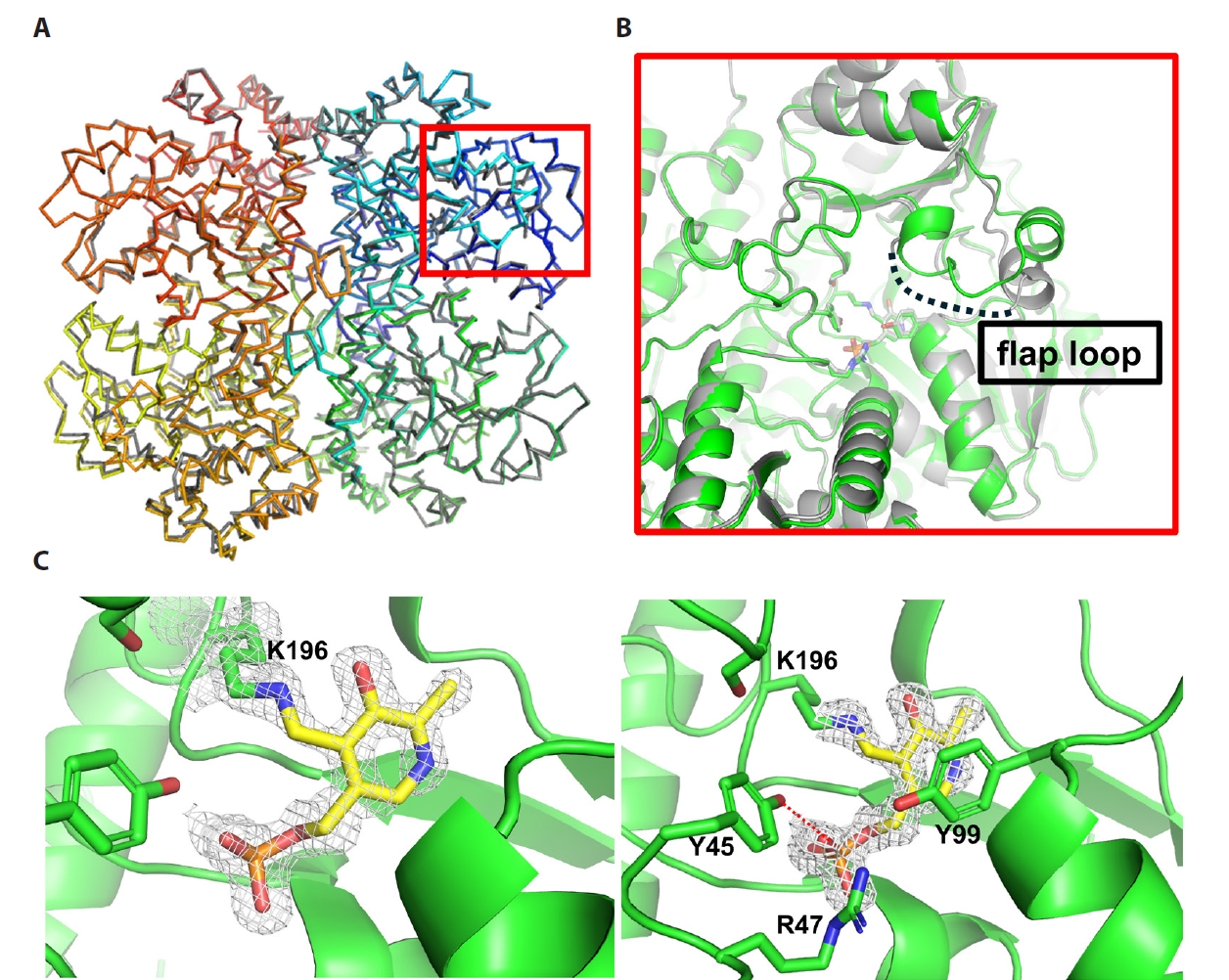
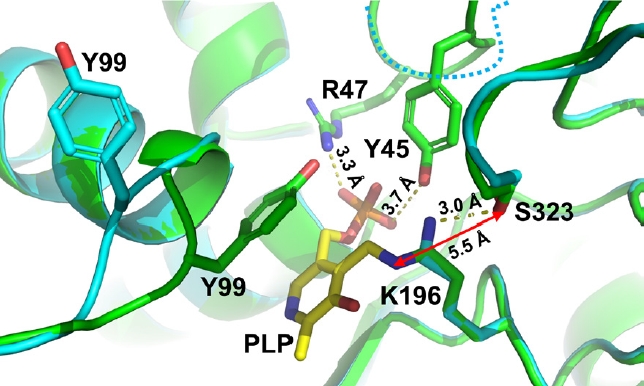
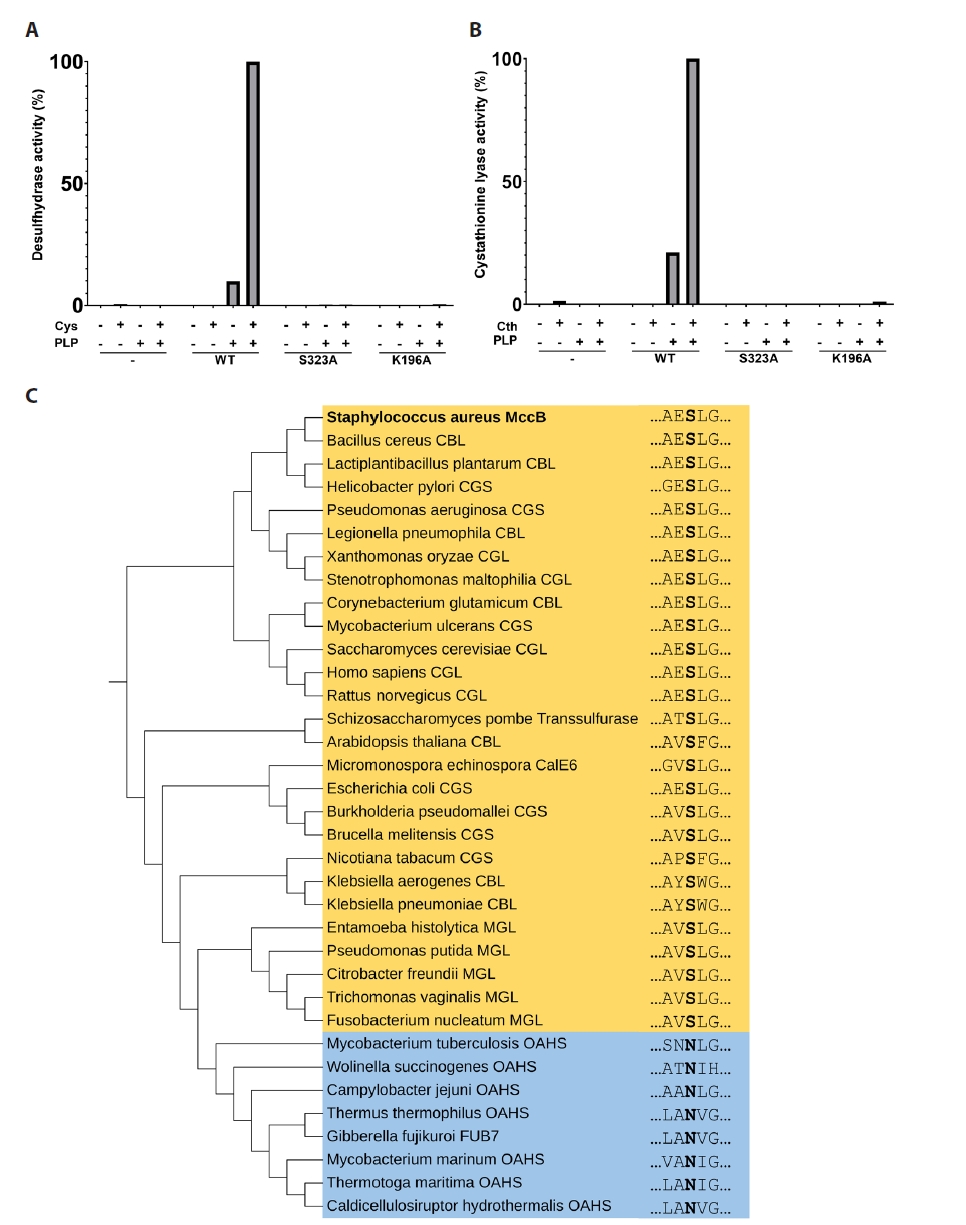
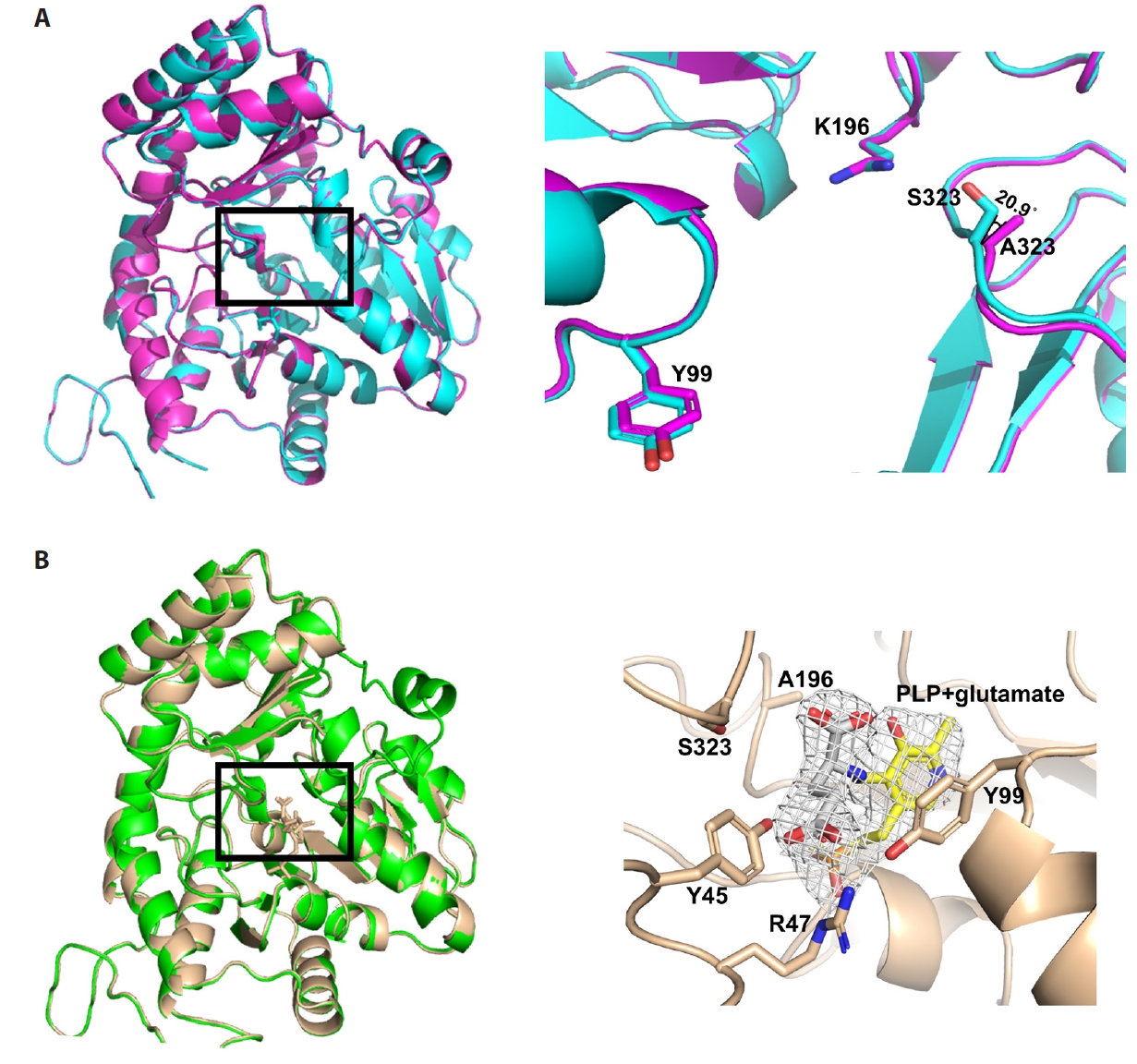

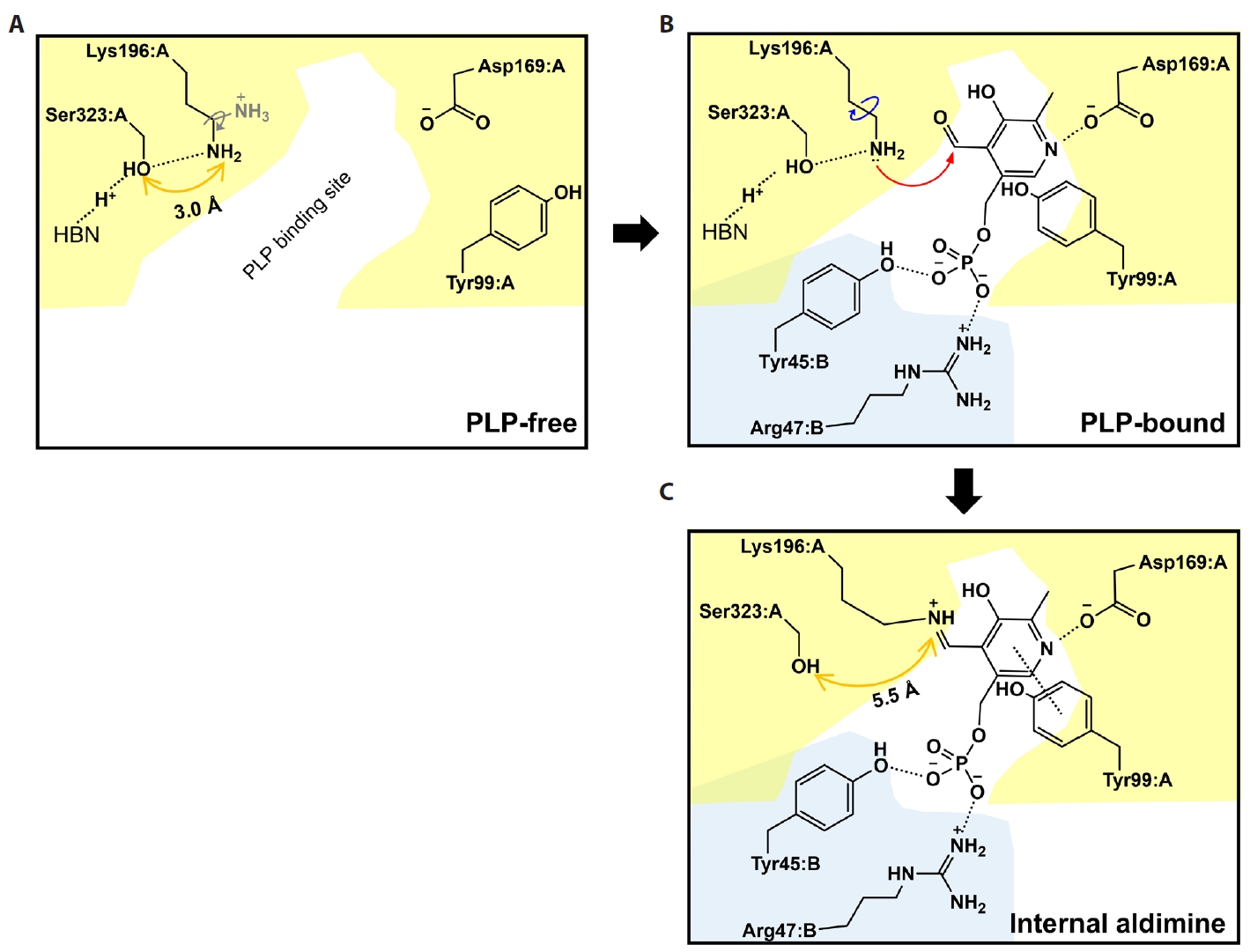
Fig. 1.
Fig. 2.
Fig. 3.
Fig. 4.
Fig. 5.
Fig. 6.
| Data collection | Wild-type, internal aldimine | S323A variant, apo-form | K196A variant, external aldimine |
|---|---|---|---|
| Beamline | PAL 5C | PAL 5C | PAL 5C |
| Wavelength (Å) | 1.00000 | 1.00000 | 1.00000 |
| Space group | I222 | I41221 | C2221 |
| a, b, c (Å) | 62.7, 80.3, 161.4 | 105.2, 105.2, 289.4 | 56.0, 154.5, 150.0 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 |
| Resolution (Å) | 50.0–1.64 (1.67–1.64) | 50.0–2.19 (2.24–2.19) | 50.0–2.5 (2.54–2.50) |
| Rpim | 0.016 (0.086) | 0.018 (0.087) | 0.022 (0.086) |
| I/σ | 6.81 | 2.54 | 5.85 |
| Completeness (%) | 97.2 (96.4) | 99.4 (99.7) | 92.0 (84.0) |
| Redundancy | 13.3 (13.3) | 15.9 (9.3) | 6.4 (3.5) |
| Resolution (Å) | 31.26–1.64 | 33.26–2.19 | 36.26–2.5 |
| No. of reflections | 48,591 | 40,789 | 20,756 |
| Rwork/Rfree | 0.1735/0.2006 | 0.1932/0.2229 | 0.2163/0.2660 |
| No. of total atoms | 3,165 | 2,813 | 5,700 |
| Wilson B-factor (Å) | 12.8 | 25.5 | 31.5 |
| Bond lengths (Å) | 0.002 | 0.006 | 0.002 |
| Bond angles (°) | 0.511 | 0.818 | 0.509 |
| Favored (%) | 98.40 | 97.5 | 96.5 |
| Allowed (%) | 1.33 | 2.5 | 3.5 |
| Outliers (%) | 0.27 | 0.0 | 0.0 |
| PDB code | 9J7Q | 9J7P | 9J7R |
Table 1.
TOP
 MSK
MSK

 ePub Link
ePub Link Cite this Article
Cite this Article







