ABSTRACT
- Alcohol consumption can lead to the accumulation of harmful metabolites, such as acetaldehyde, contributing to various adverse health effects, including hangovers and liver damage. This study presents a comprehensive genomic and functional analysis of Leuconostoc suionicum VITA-PB2, a lactic acid bacterial strain isolated from kimchi, to elucidate its role in enhancing alcohol and acetaldehyde metabolism. Genomic characterization revealed key genes encoding alcohol dehydrogenase (ADH) and aldehyde dehydrogenase (ALDH), providing insights into the metabolic capabilities of strain VITA-PB2. Phylogenomic analyses confirmed its taxonomic classification and genetic similarity to other Leuconostoc species. Functional validation through in vitro and in vivo experiments demonstrated superior ethanol and acetaldehyde decomposition abilities of strain VITA-PB2, with significant reductions in blood ethanol and acetaldehyde levels observed in rats administered with the strain. Further analysis indicated that while hepatic ADH activity did not significantly increase; however, ALDH expression was elevated. This suggests that the microbial ADH of strain VITA-PB2 contributed to ethanol breakdown, while both microbial and host ALDH facilitated acetaldehyde detoxification. These findings highlight the potential of strain VITA-PB2 as a functional probiotic for mitigating the toxic effects of alcohol consumption.
-
Keywords: Leuconostoc suionicum VITA-PB2, lactic acid bacteria (LAB), alcohol and acetaldehyde metabolism, alcohol dehydrogenase (ADH), acetaldehyde dehydrogenase (ALDH)
Introduction
Alcohol consumption remains a pervasive aspect of many cultures worldwide, but its metabolism in the human body poses substantial health risks. Upon ingestion, ethanol is primarily metabolized in the liver, where it undergoes oxidation by alcohol dehydrogenase (ADH) to form acetaldehyde—a highly reactive and toxic intermediate (Teschke, 2018). Acetaldehyde is a major contributor to the adverse effects associated with alcohol consumption, including symptoms of a hangover such as nausea, vomiting, flushing, and headaches (Erilsson, 2001). Furthermore, chronic exposure to acetaldehyde is linked to more severe conditions, such as liver cirrhosis, cardiovascular diseases, and even cancer (Penning et al., 2010; Seitz et al., 2010).
To mitigate these toxic effects, acetaldehyde is further oxidized to a less harmful and easily excreted metabolite, such as acetate via acetyl coenzyme A (acetyl-CoA) by acetaldehyde dehydrogenase (ALDH), a crucial step in detoxifying alcohol (Chang et al., 2017; Tsermpini et al., 2022) However, the efficiency of this detoxification process can vary significantly among individuals, often leading to the accumulation of acetaldehyde in the bloodstream and contributing to the severity of hangover symptoms (Erilsson, 2001; Mackus et al., 2020). Acetaldehyde, a highly reactive compound, is known to form covalent bonds with proteins, impairing DNA repair mechanisms and increasing carcinogenesis potential (Tuma et al., 1987). This interaction leads to oxidative stress and liver damage, including conditions like fatty liver and hepatic necrosis. Consequently, there is increasing interest in developing functional foods and supplements that enhance the activity of ADH and ALDH, promoting efficient alcohol metabolism and mitigating the toxic effects of alcohol consumption (Moslemi et al., 2022).
Probiotics, particularly lactic acid bacteria (LAB), have emerged as a promising approach in this regard. LAB are well-known for their beneficial effects on gut health, but recent studies have expanded their potential applications to include the enhancement of alcohol metabolism (Jung et al., 2021). Certain LAB strains, including those from the Leuconostoc genus, have demonstrated the ability to modulate alcohol metabolism by increasing the activity of ADH and ALDH (Yun et al., 2024). This effect not only accelerates the conversion of ethanol to less harmful metabolites but also reduces the concentration of acetaldehyde in the bloodstream, potentially alleviating the symptoms of alcohol-induced hangovers (Jung et al., 2023).
The efficacy of LAB in promoting alcohol metabolism has been observed in various forms, including both live and heat-killed preparations (Jung et al., 2021, 2023). For instance, studies have shown that Limosilactobacillus fermentum (formally Lactobacillus fermentum) can significantly decrease blood alcohol levels and enhance liver function by boosting ADH and ALDH activities (Jung et al., 2023). Furthermore, the use of heat-treated LAB has been found to retain these beneficial effects, suggesting that the active components responsible for this enhancement are not compromised by thermal processing (Jung et al., 2021). Given these findings, there is considerable interest in exploring other LAB strains, such as those within the Leuconostoc genus, for their potential to improve alcohol metabolism and reduce the adverse effects of alcohol consumption.
This study aims to investigate the impact of feeding a LAB strain, Leuconostoc suionicum VITA-PB2, on the degradation of alcohol and acetaldehyde in an in vitro cell system and an in vivo mouse model. Specifically, we assessed the effects of this probiotic on blood ethanol and acetaldehyde concentrations, as well as the activity of ADH and ALDH in liver tissues. A comprehensive genomic analysis was conducted to identify key genes encoding ADH and ALDH, providing fundamental insights into the metabolic capabilities of the strain. The presence of these genes highlights the mechanistic potential of VITA-PB2 to actively degrade ethanol and detoxify acetaldehyde. By elucidating the mechanisms through which Leuconostoc species influence alcohol metabolism, this research aims to establish VITA-PB2 as a promising functional ingredient in hangover relief products and other applications targeting the toxic effects of alcohol.
Materials and Methods
Preparation and isolation of the LAB strain
The kimchi sample used in this study was a traditionally fermented Korean kimchi, prepared by a local household in Daejeon, Republic of Korea, using conventional ingredients and fermentation methods. This sample was transported under 4ºC and processed for strain isolation immediately upon arrival at the laboratory. The supernatant of the kimchi sample was serially diluted in de Man Rogosa-Sharpe broth medium (MRS; Becton, Dickinson and Company, USA). Thereafter, aliquots of each serial dilution were spread on MRS agar plates, which were then incubated at 37ºC for 2 days. Colonies with distinct morphologies were picked and further purified by repeated streaking on MRS agar. The isolates were preliminarily screened based on their alcohol tolerance in vitro first. The isolates were identified through 16S rRNA gene sequencing following established protocols (Clarridge III, 2004; Johnson et al., 2019). Among the isolates, Leuconostoc suionicum VITA-PB2 and Pediococcus pentosaceus VITA-PK1 were selected for further study based on their superior ethanol and acetaldehyde metabolism. These strains were cultured in MRS broth at 37°C for 3 days and harvested by centrifugation at 8,000 × g for 15 min at 4°C. For comparative analysis, Lactobacillus brevis K2 and Lactobacillus fermentum K4 were isolated from a commercially available hangover relief product in Korea. These strains were cultured and analyzed under the same conditions as the isolates from kimchi. All experimental procedures were conducted independently and adhered to ethical standards, complying with relevant legal and regulatory requirements. Thereafter, the harvested cells were washed three times with phosphate-buffered saline (PBS), followed by the resuspension of the strain in PBS to a final concentration of 5×108 colony forming units (CFU)/100 µl and equal mixing for administration via gavage. The cell growth and density in liquid media were determined using a spectrophotometer (OD600).
Genome analysis
Genomic DNA (gDNA) was extracted from actively growing liquid cultures of the strain during the logarithmic phase. Cell pellets were obtained by centrifugation and extracted using the DNeasy PowerSoil Pro Kit (Qiagen) following the manufacturer’s protocol. The quantity and quality of the gDNA were measured using a Qubit 4 fluorometer and a DS-11 spectrophotometer (DeNovix, USA) in Bio-Health Materials Core-Facility, Jeju National University.
Long-read sequencing libraries were prepared using the Native Barcoding Kit 24 V14 (SQK-NBD114.24), and sequencing was performed on the MinION sequencing platform (Oxford Nanopore Technology) with an R.10.4.1 flow cell (FLO-MIN114). Base-calling was conducted using MinKNOW (v23.04.6), and genome assembly was performed using the wf-bacterial-genomes pipeline embedded in EPI2ME. Annotation of the assembled genome was performed with the Prokka annotation pipeline (v1.14.6) ((Seemann, 2014). Homologous genes for metabolic pathways were BLAST-searched against non-redundant protein sequences in the KEGG (Kanehisa et al., 2016) and UniRef90 (Suzek et al., 2007) databases to identify genes involved in metabolic traits. Functional assignment of the predicted genes was improved by using a set of public databases [InterPro (Paysan-Lafosse et al., 2022), Gene Ontology (Ashburner et al., 2000; Consortium et al., 2023), Pfam (Mistry et al., 2021), the Conserved Domains Database (Wang et al., 2023), TIGRFAM (Selengut et al., 2007), and EggNOG (Hernández-Plaza et al., 2023)]. Prediction of signal peptides and transmembrane helices was performed by using the web-based services SignalP (v5.0) (Almagro Armenteros et al., 2019) and TMHMM (v2.0) (Krogh et al., 2001) with default settings. Finally, the presence of antibiotic-resistance genes was assessed using ResFinder (v4.3.3) (Ferrer Florensa et al., 2022).
Phylogenomic analysis was performed using the Anvi’o phylogenomics workflow (Eren et al., 2015) and the "Bacteria_71" single-copy marker gene set included in Anvi’o. A total of 49 genes, present in the genomes of the Lactobacillaceae family obtained from the NCBI, were selected for this analysis. The tree was constructed with IQ-TREE (v2.0.3) (-m MFP -B 1000) (Nguyen et al., 2014).
Average nucleotide identity (ANIb) was calculated by pyani (v0.2.13.1) (Pritchard et al., 2016) with BLASTn alignment. The resulting percentage identity and alignment coverage between genome pairs were presented in a table. Average amino acid identity (AAI) between genomes was determined using EzAAI (v1.2.1) (Kim et al., 2021).
In vitro experiments
The alcohol tolerance of each lactic acid bacteria (LAB) strain was assessed using a modified method involving incubation in MRS broth containing 10% ethanol (v/v) (Kang et al., 2024; Yun et al., 2024). LAB cultures were initially centrifuged at 6,000 rpm for 10 min to remove the supernatant, followed by three washes with phosphate-buffered saline (PBS, pH 7.4). The resulting pellet was resuspended in MRS broth, and 99.9% EtOH (Merk Millipore) was added to reach a final ethanol concentration of 10% (v/v). The bacterial suspension was adjusted to a concentration of 109 CFU/ml and inoculated into the ethanol-containing broth at 10% (v/v). Incubation occurred at 37°C for 4 h. Alcohol tolerance was quantified by measuring the optical density at 600 nm (OD600) (A1) and comparing the results to a control culture (A0). Alcohol tolerance was calculated using the following formula:
Alcohol Tolerance (%) = (A1/A0) × 100
To evaluate the ethanol decomposition ability of LAB strains, ethanol was added to the bacterial suspensions at a final concentration of 2% (v/v). Similarly, the acetaldehyde decomposition capacity was assessed by incubating the LAB strains in MRS broth supplemented with acetaldehyde (CH3CHO) at a concentration of 8 mg per 50 ml. Each bacterial suspension (adjusted to 109 CFU/ml) was inoculated into the ethanol or acetaldehyde-containing broth at a ratio of 1:15. The bacterial cultures were incubated at 37°C for 4 h. Ethanol concentrations in the culture supernatants were measured using the Ethanol Assay Kit (Megazyme, Ireland), while acetaldehyde concentrations were determined using the Acetaldehyde Assay Kit (Megazyme, Ireland), both according to the manufacturer’s instructions. The decomposition rates were calculated by comparing the ethanol or acetaldehyde concentrations in the culture supernatant with those in a blank control, where distilled water was used in place of the bacterial suspension.
All in vitro experiments were performed in triplicate, and data were expressed as mean ± standard deviation (SD). Statistical comparisons between groups were conducted using one-way ANOVA followed by Tukey's post hoc test for multiple comparisons. Statistical significance was defined as p < 0.05.
In vivo experiments
To assess the impact of L. suionicum VITA-PB2 on ethanol and acetaldehyde metabolism, in vivo experiments were conducted using a Sprague-Dawley rat model. The study was conducted following the Laboratory Animal Act and was approved by the Institutional Animal Care and Use Committee (IACUC) of Jeju National University under protocol number approval ID # 2022-0057.
Male Sprague-Dawley rats, aged eight weeks and weighing between 250–300 g, were acquired from SAMTAKO BIO KOREA (Korea). Upon arrival, the rats were housed in a controlled environment with a temperature of 22°C ± 1°C, humidity of 50% ± 10%, and a 12-h light/dark cycle. They were acclimated for one week, during which they had unrestricted access to standard chow and water. On the final day of treatment, the rats were fasted for 18 h with only water provided. The rats were randomly divided into three groups (n = 7–8 per group): a control group that received 200 µl (2 ml/kg) of saline; a negative group that was administered with 25% ethanol (10 ml/kg) after 30 min of the experiment began, and a VITA-PB2 group that was administered 109 cells of strain VITA-PB2 by orally, followed by 25% ethanol (10 ml/kg) 30 min after the experiment began. The bacterial strain was cultured in MRS broth, harvested, and resuspended in phosphate-buffered saline (PBS) to achieve the desired concentrations. Blood samples were collected at 1, 3, and 5 h post-ethanol administration. The rats were anesthetized using isoflurane (Virbac, France) to minimize distress, and blood was collected from the jugular vein. The collected blood was centrifuged at 3,000 rpm for 15 min at 4°C, and the plasma was stored at -70°C for further analysis.
At 6 h post-ethanol administration, the rats were sacrificed, and liver tissues were excised. The liver samples were snap-frozen in liquid nitrogen and stored at -70°C for subsequent enzymatic assays. Other portions of the liver were fixed in 10% neutral-buffered formalin for histological analysis. Blood ethanol and acetaldehyde concentrations were measured using enzymatic assay kits (see above). To assess the activity of ADH and quantify ALDH in liver tissues, the samples were homogenized in cold PBS and then centrifuged at 10,000 rpm for 20 min at 4°C. The supernatant was collected, and protein concentrations were determined using the Bradford assay. The activities of ADH and the quantification of ALDH were measured using specific assay kits (ADH-Ab102533 and ALDH-RK14173).
All data were expressed as mean ± standard deviation (SD). Statistical analyses were performed using one-way ANOVA followed by Tukey's post hoc test for multiple comparisons, with a significance level set at p < 0.05. All analyses were conducted using GraphPad Prism version 8.02.263 (USA).
Result and Discussion
Phylogenetic and genomic characterization of LAB strains
Pediococcus pentosaceus VITA-PK1 and Leuconostoc suionicum VITA-PB2 are characterized as Gram-positive, facultatively anaerobic, non-motile, and non-sporulating LAB strains isolated from traditional Korean Kimchi. To investigate the phylogenetic properties and genomic characteristics of these LAB strains, we performed genome sequencing analysis. The draft genome of VITA-PK1 is composed of a single contig, with a total genome size of 1,719,447 bp with a coverage depth of 148×. The VITA-PB2 genome consists of four contigs, with a total genome size of 2,193,477 bp with a coverage depth of 100×.
Phylogenomic analysis identified strains VITA-PK1 and VITA-PB2 as Pediococcus pentosaceus and Leuconostoc suionicum, respectively (Fig. S1), with confirmation from ANI and AAI analyses (Table S1–4). Strain VITA-PB2, initially classified as Leuconostoc mesenteroides, was reclassified as Leuconostoc suionicum following its designation as a distinct species in 2017 (Jeon et al., 2017). The G+C content of P. pentosaceus VITA-PK1 (37.29%) and L. suionicum VITA-PB2 (37.11%) were consistent with the known ranges for their respective genera: Pediococcus (37–42%) and Leuconostoc (37–38%). The strains contained 1,851 and 2,209 coding sequences (CDSs), respectively.
The ANIb values for P. pentosaceus VITA-PK1 ranged from 72.5% to 99.7% compared to other members of the genus Pediococcus, with the highest value (99.7% ANIb and 90.7% coverage) observed with P. pentosaceus CECT 8330 (Table S1). The AAI values ranged from 65.3% to 99.6%, with the highest value also observed with P. pentosaceus CECT 8330 (Table S2). For L. suionicum VITA-PB2, the ANIb values ranged from 74.0% to 98.9%, with the highest value (98.9% ANIb and 87.3% coverage) observed with L. suionicum DSM 20241 (Table S3). The AAI values ranged from 70.5% to 99.1%, with the highest value observed with L. suionicum UCMA20148 (Table S4).
P. pentosaceus CECT 8330, closely related to strain VITA-PK1, is a probiotic strain known for its role in improving gut health, particularly through modulating the gut microbiota and enhancing short-chain fatty acid metabolism (Chen et al., 2021; Dong et al., 2022; Santas et al., 2015). Recent studies have also demonstrated its potential to alleviate ethanol-induced liver injury by stabilizing gut integrity and reducing inflammation markers (Jiang et al., 2020). Similarly, L. suionicum DSM 20241, closely related to strain VITA-PB2, is recognized for its role in the fermentation of foods like kimchi and its ability to support gut health (Chun et al., 2017; Jeon et al., 2017). Genomic studies indicate its capability to synthesize essential metabolites, contributing to its probiotic potential. Furthermore, related strains such as L. mesenteroides WiKim0172 have shown alcohol tolerance and the ability to enhance alcohol metabolism by increasing ADH and ALDH activity (Yun et al., 2024), suggesting the broader utility of Leuconostoc species in mitigating alcohol-induced damage.
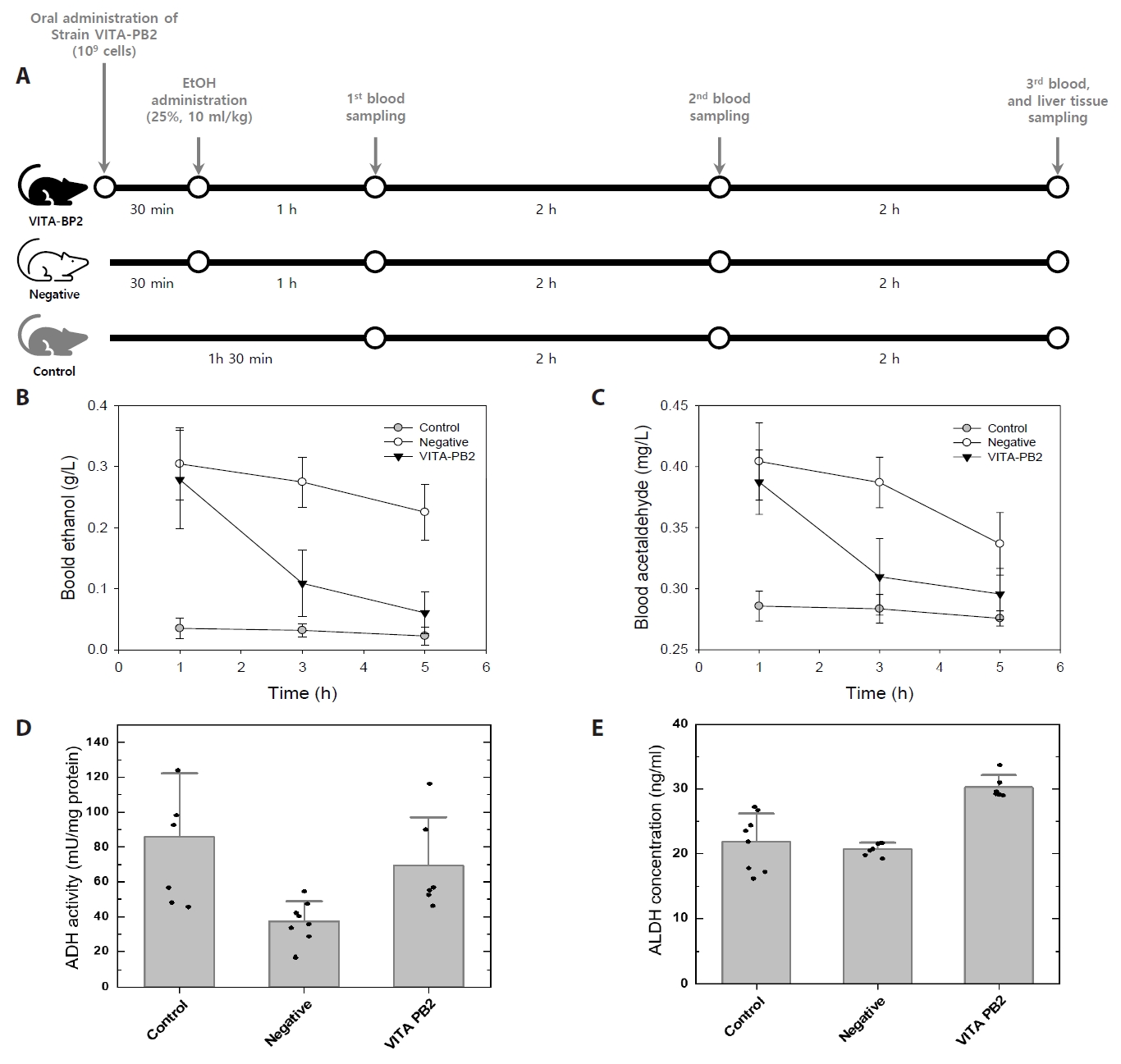
Strain VITA-PB2 employs a heterolactic fermentative pathway, converting sugars into lactic acid, ethanol, and CO₂. It utilizes metabolic pathways such as pentose phosphate, fructose and mannose, and pyruvate metabolism, but lacks a complete tricarboxylic acid (TCA) cycle (Fig. 1; Table S5 and Dataset S2). Genes for the phosphotransferase system (PTS) enable the utilization of diverse carbon sources like fructose, mannose, and sucrose, which are processed into lactate, ethanol, or acetate with CO₂ as a byproduct (Fig. 1; Dataset S1). VITA-PB2 also synthesizes polysaccharides like levanbiose and dextran (Fig. 1; Table S5 and Dataset S1), offering probiotic benefits such as pathogen inhibition and stress protection (Pramudito et al., 2024; Yokota et al., 1993). Like other Leuconostoc spp., it contains a gene set associated with bacteriocin production (Table S5 and Dataset S1).
Upon analyzing the genes involved in alcohol and acetaldehyde degradation, the VITA-PK1 genome was found to contain three ADH genes, with EC number 1.1.1.- (VITAPK1_00365, VITAPK1_00541, and VITAPK1_00710), but lacks genes encoding ALDH, as indicated in Table S1 and Dataset S1. In contrast, the VITA-PB2 genome harbors four ADH genes, along with a bifunctional acetaldehyde-CoA/alcohol dehydrogenase with EC number EC 1.2.1.10 and 1.1.1.1, as well as an ALDH gene (EC 1.2.1.3), as indicated in Table S1 and Dataset S2. This genetic composition suggests that strain VITA-PB2 has potential to metabolize ethanol and acetaldehyde effectively.
Strain VITA-PB2 lacks key genes for a complete electron transport chain (ETC) (Fig. 1; Table S5 and Dataset S1), including Complex III (EC 7.1.1.8) and catalase (EC 1.11.1.6), making it sensitive to oxygen exposure. To mitigate oxidative stress, VITA-PB2 relies on a high-affinity cytochrome bd complex (CydA and CydB) and a thiol-exporting ABC transporter (CydDC) instead of the more efficient cytochrome c oxidase (Fig. 1; Table S5 and Dataset S1). This cytochrome bd complex reduces oxygen to water with lower proton-pumping efficiency, enabling survival in low-oxygen or microaerophilic conditions, such as fermentation environments (Borisov et al., 2011). Additionally, VITA-PB2 harbors genes for NADH:quinone reductase, NAD(P)/FAD-dependent oxidoreductases, NADH-dependent oxidoreductase, NADH peroxidase, NADH oxidase and NADH-dependent flavin oxidoreductase (Fig. 1; Table S5 and Dataset S1), which facilitate NAD+ regeneration and aid in maintaining the NADH/NAD+ balance (Higuchi et al., 1999; Koike et al., 1985). These mechanisms, combined with the cytochrome bd complex, help mitigate reactive oxygen species production (Sena et al., 2024). However, the absence of Complex I and a complete TCA cycle may limit the ability of strain VIITA-PB2 to thrive in highly oxygenated environments (Shin & Han, 2015).
Alcohol metabolism in in vitro test
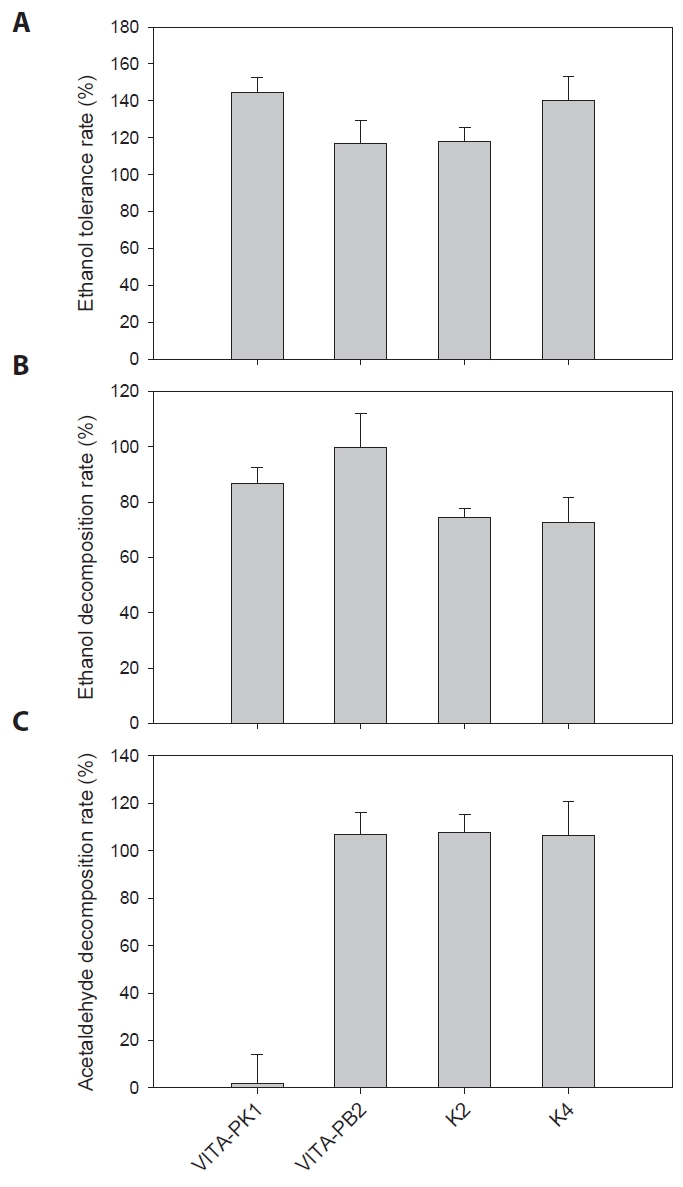
The in vitro experiments were conducted to evaluate the alcohol tolerance and the ethanol and acetaldehyde decomposition abilities of 2 isolated strains (P. pentosaceus VITA-PK1 and L. suionicum VITA-PB2) and 2 control strains (Lactobacillus brevis K2 and Lactobacillus fermentum K4). The results from the alcohol tolerance test indicated that all four LAB strains demonstrated significant resistance to ethanol, with ethanol tolerance values exceeding 100% within 4 h (Fig. 2A). This suggests that the LAB strains we tested in this study can survive and potentially function in high-alcohol environments, which is crucial for their application in fermentation processes or in managing alcohol metabolism in food products.
The ethanol decomposition test further demonstrated the metabolic capabilities of these LAB strains. As shown in Fig. 2B, strain VITA-PB2 was the most efficient, achieving an ethanol decomposition rate of ~100%. This capacity of strain VITA-PB2 to rapidly convert ethanol into acetaldehyde and subsequently into non-toxic byproducts such as acetate suggests its potential as a candidate for the development of functional foods aimed at lowering blood alcohol levels and mitigating the toxic effects of alcohol. While it is known that different bacterial strains produce varying concentrations of organic acids (Martino et al., 2022; Tagaino et al., 2019), the specific concentrations of acetate and other organic acids were not quantified in this study, leaving the precise end-products of acetaldehyde decomposition uncertain. Other tested strains also exhibited ethanol decomposition rates exceeding 70%. ADH is a group of enzymes present in many organisms that catalyze the interconversion of alcohols and aldehydes or ketones, accompanied by the reduction of nicotinamide adenine dinucleotide (NAD+) to NADH under mostly aerobic conditions (Wu et al., 2021). The presence of ADH genes in strains VITA-PB2 and VITA-PK1 likely contributes to their superior ethanol decomposition abilities, as ADH facilitates ethanol oxidation to acetaldehyde.
The acetaldehyde decomposition capabilities of the LAB strains were evaluated, and the results are presented in Fig. 2C. Strain VITA-PB2 demonstrated a remarkable ability to decompose acetaldehyde, higher than 100% decomposition within the 4-h incubation period. L. brevis K2 and L. fermentum K4 also exhibited high acetaldehyde decomposition rates, while strain VITA-PK1 showed almost no activity in acetaldehyde breakdown because of the absence of ALDH gene in the genome. The strong acetaldehyde decomposition ability of strain VITA-PB2 can be attributed to the presence of ALDH genes identified in the genome. The ability to efficiently convert acetaldehyde is a key factor in reducing the toxic burden of alcohol consumption, thereby mitigating hangover symptoms and other adverse effects associated with alcohol metabolism (Fukui et al., 1988; Konkit et al., 2015; Tagaino et al., 2019).
The ALDH enzyme plays a crucial role in detoxifying acetaldehyde during alcohol metabolism by catalyzing its oxidation to acetate through a two-step reaction that also requires NAD+ as a cofactor, with the activity of the enzyme being tightly regulated by the availability of cofactors and its structural integrity; studies have shown that ALDH forms high-order assemblies like spirosomes, which facilitate efficient substrate channeling and enzymatic turnover, especially under conditions of high acetaldehyde concentration, such as excessive alcohol consumption, thus preventing the accumulation of toxic acetaldehyde and mitigating its harmful effects, including liver damage and hangover symptoms (Fukui et al., 1988; Konkit et al., 2015; Tagaino et al., 2019).
The results of this study align with previous findings that have highlighted the importance of strain-specific capabilities in LAB for alcohol metabolism. For example, Limosilactobacillus fermentum has been shown to significantly reduce blood alcohol levels and enhance liver function by boosting ADH and ALDH activities (Kim et al., 2003). Similarly, the robust performance of Leuconostoc suionicum VITA-PB2 in both ethanol and acetaldehyde decomposition underscores the potential of certain LAB strains to serve as effective agents in functional foods or supplements designed to mitigate the toxic effects of alcohol consumption (Yun et al., 2024). The strain-specific nature of these metabolic abilities emphasizes the need for careful selection of LAB strains based on their genetic and enzymatic profiles.
Collectively, the in vitro tests demonstrate that strain VITA-PB2 is particularly effective in alcohol metabolism, exhibiting strong resistance to ethanol and remarkable capabilities in decomposing both ethanol and acetaldehyde. Therefore, while these in vitro results are promising, additional experiments were necessary to verify the alcohol degradation ability of strain VITA-PB2 in vivo, as the complexities of the biological environment can influence how effectively such mechanisms function in a living organism.
Alcohol metabolism in in vivo test
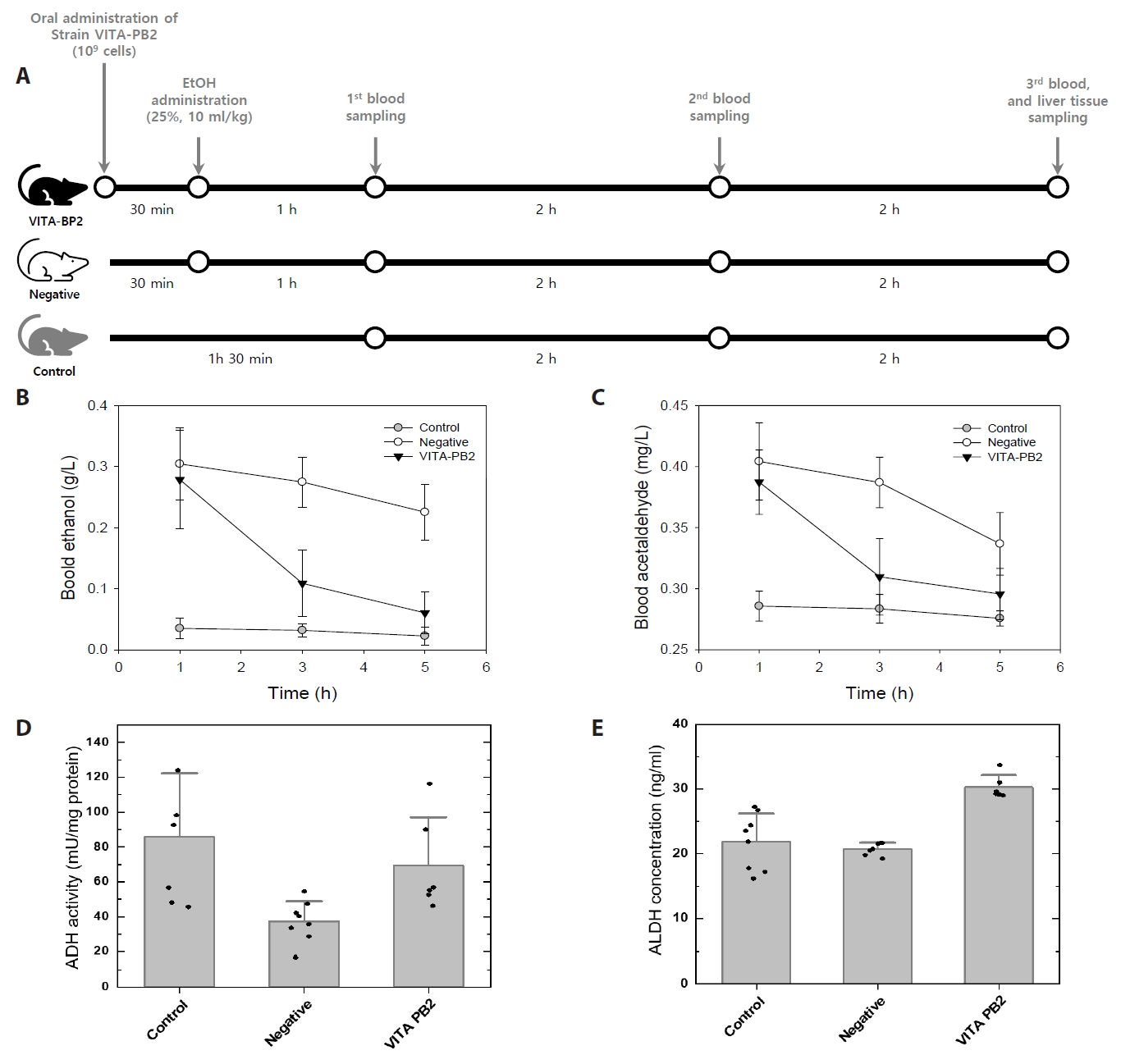
The efficacy of L. suionicum VITA-PB2 in modulating alcohol metabolism was evaluated through in vivo experiments using a Sprague-Dawley rat model (Fig. 3A). The primary focus was on assessing the impact of this LAB strain on blood ethanol and acetaldehyde concentrations, as well as the activities of ADH and ALDH in liver tissues, following ethanol administration.
Following ethanol administration, the ethanol-treated groups (VITA-PB2 and negative groups) exhibited a typical ethanol metabolism curve, with peak blood ethanol levels observed at approximately 1-h post-administration (Fig. 3B). In contrast, the groups administered with strain VITA-PB2 showed a significant reduction in blood ethanol levels compared to the control group. At the 3 h, the VITA-PB2 group showed a 68% reduction in blood ethanol concentration relative to the negative group (p < 0.05). This reduction further increased to 81% by the 5 h (p < 0.05), suggesting a potential role of VITA-PB2 in reducing blood alcohol levels.
In terms of acetaldehyde, which is a toxic by-product of ethanol metabolism (see above), the control group maintained elevated acetaldehyde levels throughout the observation period, reflecting the normal metabolic process of ethanol to acetaldehyde and its subsequent accumulation. However, in the VITA-PB2 groups, blood acetaldehyde levels were significantly lower than in the negative group: the VITA-PB2 group exhibited a 75% reduction in acetaldehyde levels at 3 h and a 67% reduction at 5 h post-ethanol administration (p < 0.05) (Fig. 3C). These results suggest that strain VITA-PB2 not only accelerates ethanol metabolism but also facilitates the rapid detoxification of acetaldehyde, converting it into acetate more efficiently.
To further understand the metabolic impacts of strain VITA-PB2 administration, the activity of ADH and expression of ALDH enzymes in liver tissues were measured. The results revealed a significantly higher activity of ADH and expression of ALDH in the VITA-PB2 groups than in the negative group (Fig. 3D & 3E). ADH activity in the VITA-PB2 group showed a non-significant reduction compared to the control group (p > 0.05), but it was 1.75-fold higher than that in the negative group (p < 0.05) (Fig. 3D). In contrast, expression of ALDH in the VITA-PB2 group was approximately 1.5-fold higher than in the control and negative groups (p < 0.05) (Fig. 3E). The ADH activity in liver tissues could decrease following excessive ethanol exposure (He et al., 2002; Tran et al., 2015). The difference in ADH activity in liver tissues between the VITA-PB2 group and the negative group, both of which were orally administered alcohol, along with the reduction in blood ethanol levels observed in Fig. 3B, suggests that the liver tissues of the VITA-PB2 group may have been less exposed to alcohol due to the intrinsic ADH activity of the administered strain and/or the gut microbiota. This study aligns with previous findings by Yun et al. (2024) and Lu et al. (2020), demonstrating that LAB strains significantly contribute to ethanol and acetaldehyde metabolism. The increase in ALDH expression observed in the VITA-PB2 group indicates that administration of LAB not only prevents the acetaldehyde accumulation within microbial cells but also enhances hepatic ALDH expression in the host. This enhancement facilitates more efficient detoxification and improved liver function (Gan et al., 2021; Kim et al., 2003).
LAB strains, such as Leuconostoc, utilize ADH in ethanol-tolerant environments to oxidize ethanol into acetaldehyde during reverse fermentation (Jung et al., 2021; Konkit et al., 2015). Meanwhile, mammalian ADH and ALDH enzymes are highly compartmentalized and tightly regulated in liver tissues (Deltour et al., 1999), LAB enzymes operate within broader fermentation networks, responding dynamically to environmental ethanol and acetaldehyde concentrations (Fukui et al., 1988; Lu et al., 2020; Salaspuro, 1997; Tagaino et al., 2019). In this study, strain VITA-PB2 demonstrated a dual role in ethanol metabolism: microbial ADH facilitated ethanol breakdown, and elevated ALDH activity efficiently reduced acetaldehyde levels in both microbial systems and host liver tissues (Fig. 3). This synergistic action accelerated ethanol clearance and mitigated acetaldehyde toxicity, aligning with previous findings that LAB administration enhances ALDH activity and improves liver function (Kim et al., 2003; Lu et al., 2020; Yun et al., 2024). The increased activity of ADH and expression of ALDH in liver tissues observed in the VITA-PB2 group provide mechanistic evidence for the reduced blood ethanol and acetaldehyde levels (Fig. 3). These results underscore the potential of strain VITA-PB2 as a functional probiotic capable of protecting against alcohol-induced liver damage and related chronic conditions, such as fatty liver disease and hepatic cirrhosis.
Conclusion
This study demonstrates the significant potential of Leuconostoc suionicum VITA-PB2 in enhancing alcohol metabolism, positioning it as a promising candidate for functional foods or supplements aimed at mitigating the toxic effects of alcohol consumption. Both in vitro and in vivo experiments confirmed the efficacy of strain in reducing blood ethanol and acetaldehyde levels, highlighting its ability to accelerate alcohol clearance and detoxify harmful by-products. The increased expression of ALDH observed in the VITA-PB2-treated groups, along with its intrinsic ADH activity, suggests that the strain not only assists in ethanol breakdown but also enhances the detoxification of host pathways. This dual mechanism makes strain VITA-PB2 an effective probiotic for improving alcohol metabolism and reducing symptoms such as hangovers. Given its superior performance, particularly in decomposing acetaldehyde, strain VITA-PB2 offers a novel approach to managing the adverse effects of alcohol consumption, such as hangovers and alcohol-induced liver damage. These findings suggest that regular administration of strain VITA-PB2 could provide long-term protective effects, potentially preventing chronic alcohol-related conditions such as fatty liver disease and hepatic cirrhosis. Overall, the strain-specific advantages of the LAB strain emphasize the need for targeted probiotic applications in alcohol metabolism and lay the groundwork for future studies focusing on its broader health benefits.
Acknowledgments
This research was supported by the Research Institute for Basic Sciences (RIBS) of Jeju National University (2019R1A6A1A10072987), funded by the Ministry of Education, and the National Research Foundation of Korea (NRF-2021R1C1C1008303 and NRF-2022R1A4A503144711), funded by MSIT.
Conflict of Interest
The strains used in this study, including Leuconostoc suionicum VITA-PB2, are currently under research and development, and no commercial product is available at this stage. The authors declare no conflicts of interest regarding the conduct of this research or potential future applications.
Ethical Statements
This article does not contain any studies with human participants by any authors.
Supplementary Information
The online version contains supplementary material available at https://doi.org/10.71150/jm.2410026.
Table S1.
The pairwise comparison of average nucleotide identity (ANI) values for the genomes of Pediococcus pentosaceus VITA-PK1 and reltated strains.
jm-2410026-Supplementary-Table-S1.pdf
Table S2.
The pairwise comparison of Average Amino Acid Identity (AAI) values for the genomes of Pediococcus pentosaceus VITA-PK1 and related strains.
jm-2410026-Supplementary-Table-S2.pdf
Table S3.
The pairwise comparison of average nucleotide identity (ANI) values for the genomes of Leuconostoc suionicum VITA-PB2 and reltated strains.
jm-2410026-Supplementary-Table-S3.pdf
Table S4.
The pairwise comparison of Average Amino Acid Identity (AAI) values for the genomes of Leuconostoc suionicum VITA-PB2 and related strains.
jm-2410026-Supplementary-Table-S4.pdf
Fig. S1.
Phylogenomic tree and average nucleotide and amino acid identity (ANI and AAI) matrices for the genomes of Pediococcus pentosaceus VITA-PK1 and Leuconostoc suionicum VITA-PB2 strains, along with related species within the family Lactobacillaceae. Phylogenomic tree constructed based on 49 genes from the "Bacteria_71" single-copy marker gene set using Anvi’o’s phylogenomic workflow. The maximum-likelihood phylogeny was inferred using IQ-TREE (options: -B 1000 -m LG+F+I+R6). Black circles at the nodes indicate bootstrap support of 70% or higher.
jm-2410026-Supplementary-Fig-S1.pdf
Fig. 1.Genomic metabolic pathways identified in Leuconostoc suionicum VITA-PB2. Enzymes involved in the aerobic metabolic pathways for ethanol and acetaldehyde consumption are highlighted in red. The pathway between ethanol and acetate is referenced by Tagaino et al. (2019), and Fukui et al. (1988), and the KEGG pathway. Graphical representation of metabolic pathways for carbon utilization, with focus on pentose phosphate, fructose, and mannose pathways. Gene presence for phosphotransferase system (PTS), enabling carbon source utilization and fermentation processes. UDP: uridine diphosphate.
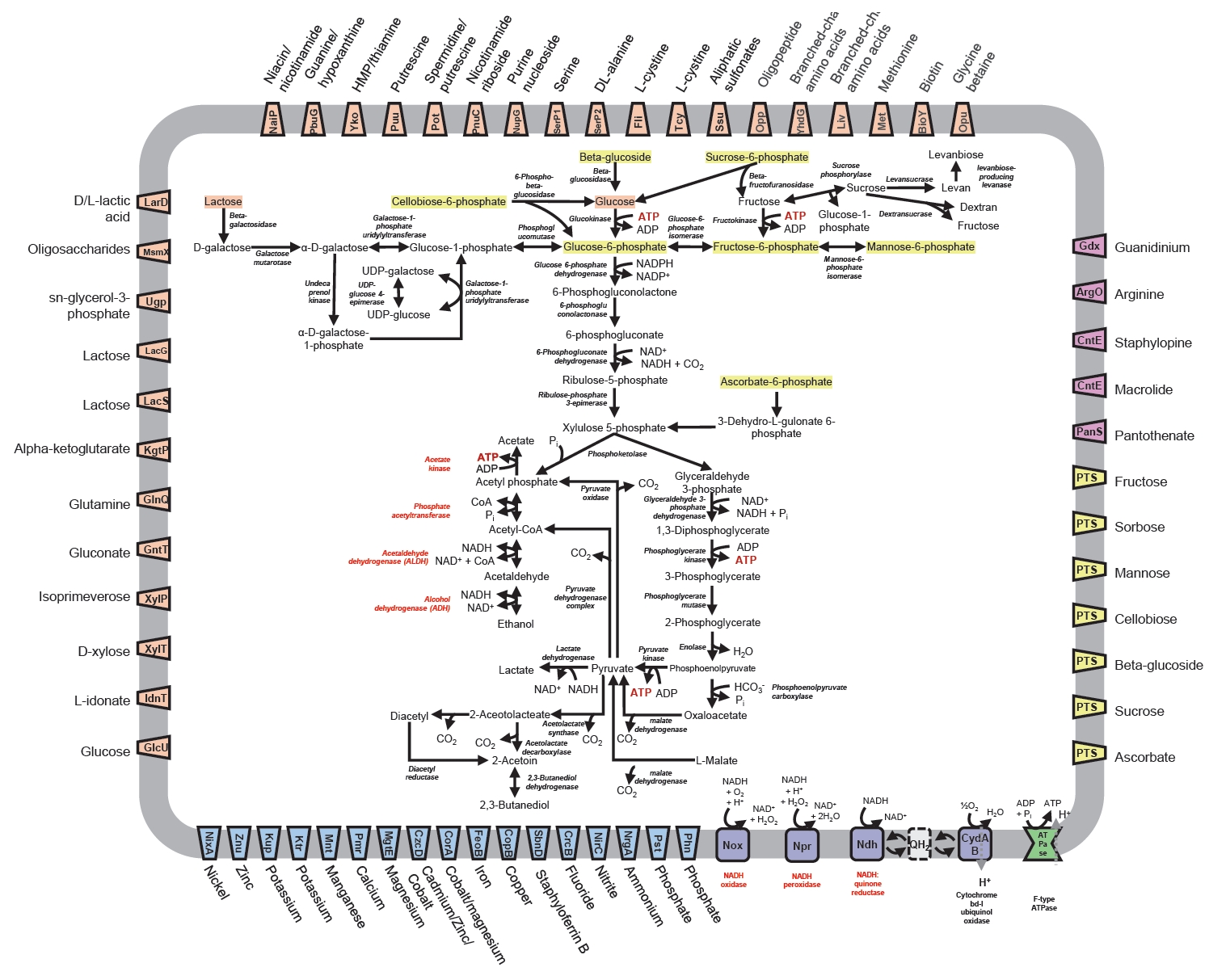
Fig. 2.Ethanol and acetaldehyde decomposition abilities of LAB strains in vitro. (A) Alcohol tolerance levels of four LAB strains, including strain VITA-PB2, after incubation in ethanol-containing media. (B) Ethanol decomposition rates of the tested strains, with strain VITA-PB2 exhibiting the highest rate of ethanol breakdown. (C) Acetaldehyde decomposition rates show the strong capability of strain VITA-PB2 to detoxify acetaldehyde compared to control strains. Values are presented as the mean ± standard deviation from triplicate experiments.
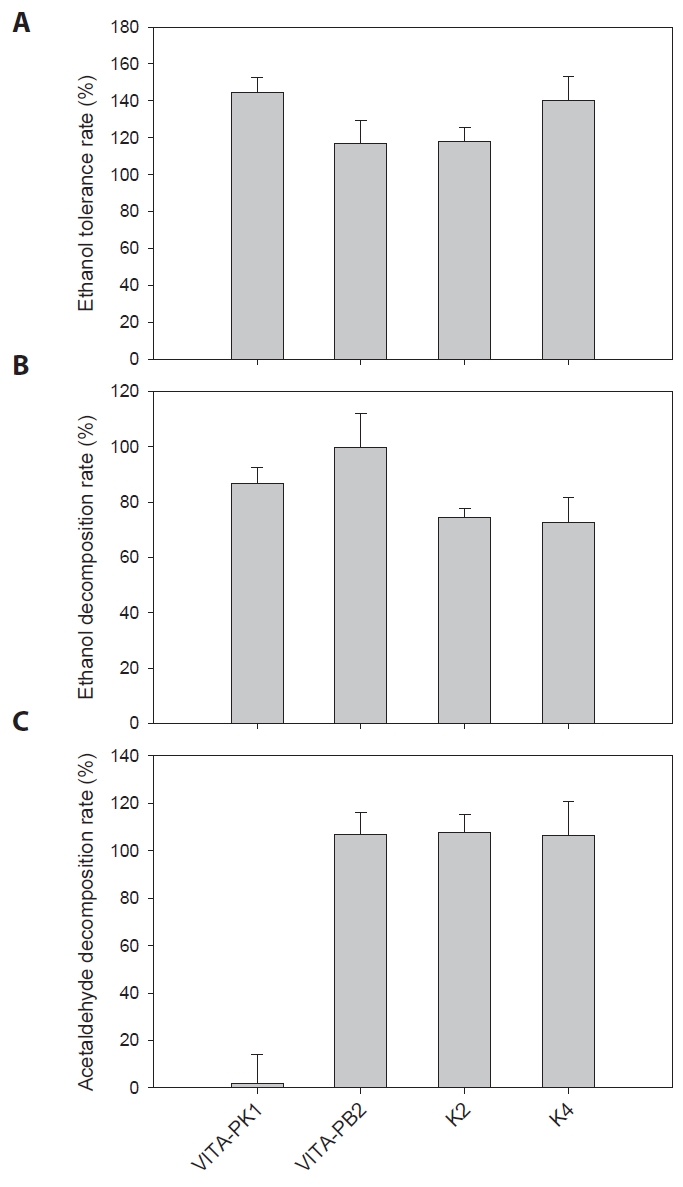
Fig. 3.In vivo effects of Leuconostoc suionicum VITA-PB2 on ethanol and acetaldehyde metabolism in rats. (A) Experimental design for in vivo effects of Leuconostoc suionicum VITA-PB2 on alcohol metabolism in rats. The schematic illustrates the experimental timeline, including the administration of VITA-PB2 followed by ethanol treatment. Blood samples were collected at 1, 3, and 5 h post-ethanol administration, and liver tissue harvested at the 5 h mark. The experimental groups were defined as follows: control, untreated with VITA-PB2 and ethanol; negative, treated with ethanol but not VITA-PB2; VITA-PB2, treated with VITA-PB2 and ethanol. (B) Blood ethanol and (C) Blood acetaldehyde concentrations; (D) Hepatic alcohol dehydrogenase (ADH) activity and (E) aldehyde dehydrogenase (ALDH) expression were measured from liver samples collected after 5 hhour. Values are presented as the mean ± standard deviation from triplicate experiments. Significant differences between groups were determined by a t-test (*p < 0.05). A higher p-value of 0.05 was considered not significant.
References
- Almagro Armenteros JJ, Tsirigos KD, Sønderby CK, Petersen TN, Winther O, et al. 2019. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat Biotechnol. 37(4): 420–423. ArticlePubMedPDF
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, et al. 2000. Gene Ontology: tool for the unification of biology. Nat Genet. 25(1): 25–29. ArticlePubMedPMCPDF
- Borisov VB, Gennis RB, Hemp J, Berkhobsky MI. 2011. The cytochrome bd respiratory oxygen reductases. Biochim Biophys Acta. 1807(11): 1398–1413. ArticlePubMedPMC
- Chang JS, Hsiao JR, Chen CH. 2017. ALDH2 polymorphism and alcohol-related cancers in Asians: a public health perspective. J Biomed Sci. 24(19): 1–10. ArticlePubMedPMCPDF
- Chen K, Liu C, Li H, Lei Y, Zeng C, et al. 2021. Infantile colic treated with Bifidobacterium longum CECT7894 and Pediococcus pentosaceus CECT8330: a randomized, double-blind, placebo-controlled trial. Front Pediatr. 9: 635176.ArticlePubMedPMC
- Chun BH, Lee SH, Jeon HH, Kim DW, Jeon CO. 2017. Complete genome sequence of Leuconostoc suionicum DSM 20241T provides insights into its functional and metabolic feature. Stand Genomic Sci. 12: 38.ArticlePubMedPMC
- Clarridge III JE. 2004. Impact of 16S rRNA gene sequence analysis for identification of bacteria on clinical microbiology and infectious diseases. Clin Microbiol Rev. 17(4): 840–862. ArticlePubMedPMCPDF
- Consortium TGO, Aleksander SA, Balhoff J, Carbon S, Cherry JM, et al. 2023. The gene ontology knowledgebase in 2023. Genetics. 224(1): iyad031.PubMedPMC
- Deltour L, Folio MH, Duester G. 1999. Metabolic deficiencies in alcohol dehydrogenase Adh1, Adh3, and Adh4 null mutant mice. Overlapping roles of Adh1 and Adh4 in ethanol clearance and metabolism of retinol to retinoic acid. J Biol Chem. 274(24): 16796–16801. ArticlePubMed
- Dong F, Xiao F, Li X, Li Y, Wang X, et al. 2022. Pediococcus pentosaceus CECT 8330 protects DSS-induced colitis and regulates the intestinal microbiota and immune responses in mice. J Transl Med. 20: 33.ArticlePubMedPMCPDF
- Eren AM, Esen OC, Quince C, Vineis JH, Morrison HG, et al. 2015. Anvi’o: an advanced analysis and visualization platform for ‘omics data. PeerJ. 3: e1319. ArticlePubMedPMCPDF
- Erilsson CJP. 2001. The role of acetaldehyde in the actions of alcohol (update 2000). Alcohol Clin Exp Res. 25(5): 15S–32S. ArticlePubMed
- Ferrer Florensa A, Sommer Kaas R, Lanken Conradsen Clausen PT, Aytan-Aktug D, Aarestrup FM. 2022. ResFinder-an open online resource for identification of antimicrobial resistance genes in next-generation sequencing data and prediction of phenotypes from genotypes. Microb Genom. 8(1): 000748.ArticlePubMedPMC
- Fukui K, Kato K, Kodama T, Ohta H, Shimamoto T, et al. 1988. Kinetic study of a change in intracellular ATP level associated with aerobic catabolism of ethanol by Streptococcus mutans. J Bacteriol. 170(10): 4589–4593. ArticlePubMedPMCPDF
- Gan Y, Tong J, Zhou X, Long X, Pan Y, et al. 2021. Hepatoprotective effect of Lactobacillus plantarum HFY09 on ethanol-induced liver injury in mice. Frint Nutr. 8: 684588.ArticlePubMedPMC
- He L, Ronins MJJ, Badger TM. 2002. Ethanol Induction of cass I acohol dhydrogenase epression in the rat occurs through alterations in CCAAT/Enhancer binding β and γ. J Biol Chem. 277(46): 43572–43577. PubMed
- Hernández-Plaza A, Szklarczyk D, Botas J, Cantalapiedra CP, Giner-Lamia J, et al. 2023. eggNOG 6.0: enabling comparative genomics across 12535 organisms. Nucleic Acids Res. 51(D1): 389–394.
- Higuchi M, Yamamoto Y, Poole LB, Shimada M, Sato Y, et al. 1999. Functions of two types of NADH oxidases in energy metabolism and oxidative stress of Streptococcus mutans. J Bacteriol. 181(19): 5940–5947. ArticlePubMedPMCPDF
- Jeon HH, Kim KH, Chun BH, Ryu BH, Han NS, et al. 2017. A proposal of Leuconostoc mesenteroides subsp. jonggajibkimchii subsp. nov. and reclassification of Leuconostoc mesenteroides subsp. suionicum (Gu et al., 2012) as Leuconostoc suionicum sp. nov. based on complete genome sequences. Int J Syst Evol Microbiol. 67(7): 2225–2230. ArticlePubMed
- Jiang XW, Li YT, Ye JZ, Lv LX, Yang LY, et al. 2020. New strain of Pediococcus pentosaceus alleviates ethanol-induced liver injury by modulating the gut microbiota and short-chain fatty acid metabolism. World J Gastroenterol. 26(40): 6224–6240. ArticlePubMedPMC
- Johnson JS, Spakowicz DJ, Hong BY, Petersen LM, Demkowicz P, et al. 2019. Evaluation of 16S rRNA gene sequencing for species and strain-level microbiome analysis. Nat Commun. 10: 5029.ArticlePubMedPMCPDF
- Jung SJ, Hwang JH, Park EO, Lee SO, Chung YJ, et al. 2021. Regulation of alcohol and acetaldehyde metabolism by a mixture of Lactobacillus and Bifidobacterium species in human. Nutrients. 13(6): 1875.ArticlePubMedPMC
- Jung SH, Lee YH, Lee EK, Park SD, Shim JJ, et al. 2023. Effects of plant-based extract mixture on alcohol metabolism and hangover improvement in humans: a randomized, double-blind, aralleled, placebo-controlled clinical trial. J Clin Med. 12(16): 5244.PubMedPMC
- Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M. 2016. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 44(D1): 457–462. ArticlePubMedPMC
- Kang S, Long J, Park MS, Ji GE, Ju Y, et al. 2024. Investigating human-derived lactic acid bacteria for alcohol resistance. Microb Cell Fact. 23(1): 118.ArticlePubMedPMCPDF
- Kim JH, Kim HJ, Son JH, Chun HN, Yang SJ, et al. 2003. Effect of Lactobacillus fermentum MG590 on alcohol metabolism and liver function in rats. J Microbiol Biotechnol. 13(6): 919–925.PDF
- Kim D, Park S, Chun J. 2021. Introducing EzAAI: a pipeline for high throughput calculations of prokaryotic average amino acid identity. J Microbiol. 59(5): 476–480.ArticlePubMedPDF
- Koike K, Kobayashi T, Ito S, Saitoh M. 1985. Purification and characterization of NADH oxidase from a strain of Leuconostoc mesenteroides. J Biochem. 97(5): 1279–1288. ArticlePubMed
- Konkit M, Choi WJ, Kim W. 2015. Aldehyde dehydrogenase activity in Lactococcus chungangensis: application in cream cheese to reduce aldehyde in alcohol metabolism. J Dairy Sci. 99(3): 1755–1761. ArticlePubMed
- Krogh A, Larsson B, von Heijne G, Sonnhammer ELL. 2001. Predicting transmembran protein topology with a hidden markov model: application to complete genomes. J Mol Biol. 305(3): 567–580. ArticlePubMed
- Lu J, Zhu X, Zhang C, Lu F, Lu Z, et al. 2020. Co-expression of alcohol dehydrogenase and aldehyde dehydrogenase in Bacillus subtilis for alcohol detoxification. Food Chem Toxicol. 135: 110890.ArticlePubMed
- Mackus M, van de Loo AJ, Garssen J, Kraneveld AD, Scholey A, et al. 2020. The role of alcohol metabolism in the pathology of alcohol hangover. J Clin Med. 9(11): 3421.ArticlePubMedPMC
- Martino C, Parker SJ, Zaramela LS, Gao B, Embree M, et al. 2022. Acetate reprograms gut microbiota during alcohol consumption. Nat Commun. 13: 4630.ArticlePubMedPMCPDF
- Mistry J, Chuguransky S, Williams L, Qureshi M, Salazar GA, et al. 2021. Pfam: the protein families database in 2021. Nucleic Acids Res. 49(D1): D412–D419. ArticlePubMedPMCPDF
- Moslemi M, Jannat B, Mahmoudzadeh M, Ghasemlou M, Abedi AS. 2022. Detoxification activity of bioactive food compounds against ethanol-induced injuries and hangover symptoms: A review. Food Sci Nutr. 11(9): 5028–5040. ArticlePubMedPMC
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2014. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1): 268–274. ArticlePubMedPMC
- Paysan-Lafosse T, Blum M, Chuguransky S, Grego T, Lázaro Pinto B, et al. 2022. InterPro in 2022. Nucleic Acids Res. 51(D1): D418–D427. ArticlePubMedPMCPDF
- Penning R, van Nuland M, Fliervoet KAL, Olivier B, Verster JC. 2010. The pathology of alcohol hangover. Curr Drug Abuse Rev. 3(2): 68–75. ArticlePubMed
- Pramudito TE, Desai K, Voigt C, Smid EJ, Schols HA. 2024. Dextran and levan exopolysaccharides from tempeh-associated lactic acid bacteria with bioactivity against enterotoxigenic Escherichia coli (ETEC). Carbohydr Polym. 328: 121700.ArticlePubMed
- Pritchard L, Glover RH, Humphris S, Elphinstone JG, Toth IK. 2016. Genomics and taxonomy in diagnostics for food security: soft-rotting enterobacterial plant pathogens. Anal Methods. 8: 12–24.ArticlePDF
- Salaspuro M. 1997. Microbial metabolism of ethanol and acetaldehyde and clinical consequences. Addict Biol. 2(1): 35–46. ArticlePubMedPDF
- Santas J, Fuentes MC, Tormo R, Guayta-Escolies R, Lázaro E, et al. 2015. Pediococcus pentosaceus CECT 8330 and Bifidobacterium longum CECT 7894 show a trend towards lowering infantile excessive crying syndrome in a pilot clinical trial. Int J Pharm Bio Sci. 6(2): 458–466.PDF
- Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 30(14): 2068–2069. ArticlePubMedPDF
- Seitz HK, Stickel F. 2010. Acetaldehyde as an underestimated risk factor for cancer development: role of genetics in ethanol metabolism. Genes Nutr. 5(2): 121–128. ArticlePubMedPMCPDF
- Selengut JD, Haft DH, Davidsen T, Ganapathy A, Gwinn-Giglio M, et al. 2007. TIGRFAMs and genome properties: tools for the assignment of molecular function and biological process in prokaryotic genomes. Nucleic Acids Res. 35: D260–264. ArticlePubMedPMC
- Sena FV, Sousa FM, Pereira AR, Catarino T, Cabrita EJ, et al. 2024. The two alternative NADH:quinone oxidoreductases from Staphylococcus aureus: two players with different molecular and cellular roles. Microbiol Spectr. 12(8): e0415223. ArticlePubMedPMCPDF
- Shin SY, Han NS. 2015. In Beneficial Microorganisms in Food and Nutraceuticals. Microbiology Monographs, Vol. 27, pp. 111–132. Springer Nature.PDF
- Suzek BE, Huang H, McGarvey P, Mazumder R, Wu CH. 2007. UniRef: comprehensive and non-redundant UniProt reference clusters. Bioinformatics. 23(10): 1282–1288. ArticlePubMedPDF
- Tagaino R, Washio J, Abiko Y, Tanda N, Sasaki K, et al. 2019. Metabolic property of acetaldehyde production from ethanol and glucose by oral Streptococcus and Neisseria. Sci Rep. 9(1): 10446.ArticlePubMedPMCPDF
- Teschke R. 2018. Alcoholic liver disease: alcohol metabolism, cascade of molecular mechanisms, cellular targets, and clinical aspects. Biomedicines. 6(4): 106.ArticlePubMedPMC
- Tran S, Nowicki M, Chatterjee D, Gerlai R. 2015. Acute and chronic ethanol exposure differentially alters alcohol dehydrogenase and aldehyde dehydrogenase activity in the zebrafish liver. Prog Neuropsychopharmacol Biol Psychiatry. 56: 221–226. ArticlePubMed
- Tsermpini EE, Plemenitaš Ilješ A, Dolžan V. 2022. Alcohol-induced oxidative stress and the role of antioxidants in alcohol use disorder: A systematic review. Antioxidants (Basel). 11(7): 1374.ArticlePubMedPMC
- Tuma DJ, Newman MR, Donohue TMJ, Sorrell MF. 1987. Covalent binding of acetaldehyde to proteins: Participation of lysine residues. Alcohol Clin Exp Res. 11(6): 579–584. ArticlePubMed
- Wang J, Chitsaz F, Derbyshire MK, Gonzales NR, Gwadz M, et al. 2023. The conserved domain database in 2023. Nucleic Acids Res. 51(D1): D384–388. ArticlePubMedPMCPDF
- Wu YY, Lee YS, Liu YL, Hsu WC, Ho WM, et al. 2021. Association study of alcohol dehydrogenase and aldehyde dehydrogenase polymorphism with alzheimer disease in the taiwanese population. Front Neurosci. 15: 625885.ArticlePubMedPMC
- Yokota A, Kondo K, Nakagawa M, Kojima I, Tomita F. 1993. Production of levanbiose by a levan-degrading enzyme from Streptomyces exfoliatus F3-2. Biosci Biotechnol Biochem. 57(5): 745–749. Article
- Yun M, Jo HE, Kim N, Park HK, Jang YS, et al. 2024. Oral administration of alcohol-tolerant lactic acid bacteria alleviates blood alcohol concentration and ethanol-induced liver damage in rodents. J Microbiol Biotechnol. 34(4): 838–845. ArticlePubMedPMC
Citations
Citations to this article as recorded by

- Microorganisms: The Key Regulators of Wine Quality
Hechao Zhao, Shiyuan Liu, Lixian Zhu, Yanhua Wang
Comprehensive Reviews in Food Science and Food Safety.2025;[Epub] CrossRef - Efficacy of Probiotic VITA-PB2 from Fermented Foods on Alcohol Consumption and Hangover Symptoms: A Randomized, Double-Blind, Placebo-Controlled Trial
Chaodeng Mo, Johny Bajgai, Md. Habibur Rahman, Sofian Abdul-Nasir, Hui Ma, Thu Thao Pham, Haiyang Zhang, Buchan Cao, Seong Hoon Goh, Bomi Kim, Hongik Kim, Min Kyeong Seol, Young Geon Yu, Cheol-Su Kim, Kyu-Jae Lee, Seung-Taek Lim
Nutrients.2025; 17(14): 2276. CrossRef




 MSK
MSK

 ePub Link
ePub Link Cite this Article
Cite this Article




