ABSTRACT
- Antarctic fungi can effectively adapt to extreme environments, which leads to the production of unique bioactive compounds. Studies on the discovery of fungi in the diverse environments of Antarctica and their potential applications are increasing, yet remain limited. In this study, fungi were isolated from various substrates on the Fildes Peninsula in Antarctica and screened for their antibiosis activity against two significant plant pathogenic fungi, Botrytis cinerea and Fusarium culmorum. Phylogenetic analysis using multiple genetic markers revealed that the isolated Antarctic fungal strains are diverse, some of which are novel, emphasizing the underexplored biodiversity of Antarctic fungi. These findings suggest that these fungi have potential for the development of new antifungal agents that can be applied in agriculture to manage fungal plant pathogens. Furthermore, the antibiosis activities of the isolated Antarctic fungi were evaluated using a dual-culture assay. The results indicated that several strains from the genera Cyathicula, Penicillium, and Pseudeurotium significantly inhibited pathogen growth, with Penicillium pancosmium showing the highest inhibitory activity against Botrytis cinerea. Similarly, Aspergillus and Tolypocladium strains exhibited strong antagonistic effects against Fusarium culmorum. This study enhances our understanding of Antarctic fungal diversity and highlights its potential for biotechnological applications.
-
Keywords: Antarctic fungi, antibiosis, Botrytis cinerea, Fusarium culmorum
Introduction
Fungi are among the largest groups of organisms and thrive in diverse environments, where they occupy multiple ecological niches and play several roles, including saprotrophic, pathogenic, and symbiotic roles, making them essential ecosystem components (Kendrick, 2011; Naranjo-Ortiz and Gabaldón, 2019). Globally, their biomass accounts for approximately 12 gigatons of carbon (Bar‐On et al., 2018). Consequently, they exhibited a distinctive metabolic plasticity that enables rapid adaptation and survival through the biosynthesis of various natural products (Bhattarai et al., 2021; Gholami-Shabani et al., 2019). Fungi-derived natural products are pharmaceutically prolific and have been developed for several important biological applications, ranging from highly potent toxins to approved drugs (Aly et al., 2011; Rastegari et al., 2020; Schueffler and Anke, 2014; Vicente et al., 2003).
Antarctica represents one of the most extreme environments on Earth for the existence of life. This ecosystem exhibits high-stress conditions, including low temperatures, sporadic and limited nutrient availability, high aridity, and elevated ultraviolet radiation levels. Antarctic fungi must adapt to survive under these highly demanding conditions (Hassan et al., 2016). These adaptations result from modifications in gene expression and secondary metabolite biosynthesis, forming biologically relevant chemical spaces that allow them to survive efficiently in Antarctica (Varrella et al., 2021; Zucconi et al., 2020).
Studies on fungi in Antarctic ecosystems are limited. However, many studies on Antarctic fungi have explored the diversity and potential applications of culturable fungi from various Antarctic environments (González et al., 2020; Varrella et al., 2021). Despite these efforts, many studies on fungal diversity in Antarctica rely primarily on the internal transcribed spacer (ITS) region, a universal fungal genetic marker (Schoch et al., 2012), which often leads to inaccurate identification (Dupuis et al., 2012; Kiss, 2012). This difficulty in species identification limits our understanding of fungal biology and its potential applications.
Most studies on Antarctic fungi have focused primarily on the characteristics of secondary metabolites, including novel metabolite production and antibacterial properties (Ordóñez-Enireb et al., 2022; Shi et al., 2022; Vieira et al., 2018). However, their potential use, particularly in combating plant pathogens, remains undetermined. Recent findings regarding natural compounds that capable of inhibiting plant diseases have generated renewed interest (Kim and Hwang, 2007; Vinale et al., 2014; Wang et al., 2023). Therefore, Antarctic fungi may be promising candidates with hidden and remarkable capabilities.
Botrytis cinerea and Fusarium culmorum are representative plant pathogenic fungi that cause significant economic losses to agriculture. Botrytis cinerea, responsible for grey mold, is a highly destructive pathogen and is estimated to cause nearly $100 billion in annual agricultural losses (Dwivedi et al., 2024; Roca-Couso et al., 2021). The destructive nature of this fungus ranks second among scientifically and economically relevant pathogenic fungi (Dean et al., 2012). In Chile, B. cinerea affects grapes by reducing both yield and quality during ripening. Integrated management practices, including cultural, chemical, and biological methods, are crucial for controlling this pathogen in Chilean vineyards under temperate and humid conditions (Herrera-Défaz et al., 2023; Latorre et al., 2015).
Fusarium culmorum, on the other hand, affects cereals, such as wheat and barley, causing Fusarium head blight, which reduces grain yield and quality. The presence of this pathogen in Chile is significant, especially in humid areas where it can produce mycotoxins such as deoxynivalenol, posing additional food safety concerns (Scherm et al., 2013). Chemical strategies through fungicides are currently the most widely used methods for controlling infections. Both B. cinerea and F. culmorum have developed resistance to several conventional fungicides (Yin et al., 2023), causing substantial agricultural damage worldwide (Hahn, 2014). Therefore, discovering new natural molecules with high efficiency in controlling plant pathogenic fungal growth is of vital importance to the agricultural sector.
During the Antarctic expedition (ECA59) funded by the Chilean Antarctic Institute, we collected several environmental samples, including soil, lichens, plants, and snow, from the Fildes Peninsula, Antarctica. We isolated 97 fungal strains and examined their diversity and antibiosis ability against two plant pathogens. Through phylogenetic analysis using multi-genetic markers (ITS, LSU, ACT, RPB2, TEF1, and TUB) specific to each taxonomic group, we elucidated species diversity with considerable accuracy. Using a dual-culture assay approach, we evaluated the antibiosis potential of all Antarctic fungal strains against B. cinerea and F. culmorum. Several strains from the genera Aspergillus, Cyathicula, Penicillium, Pseudeurotium, Pseudogymnoascus, and Tolypocladium showed a remarkable capacity to control the growth of these phytopathogens. Thus, our study offers comprehensive insights into the diversity of culturable fungi in Antarctica and their potential for antibiosis. This study will broaden the understanding of Antarctic fungi and establish groundwork for future research.
Materials and Methods
Sample collection and processing
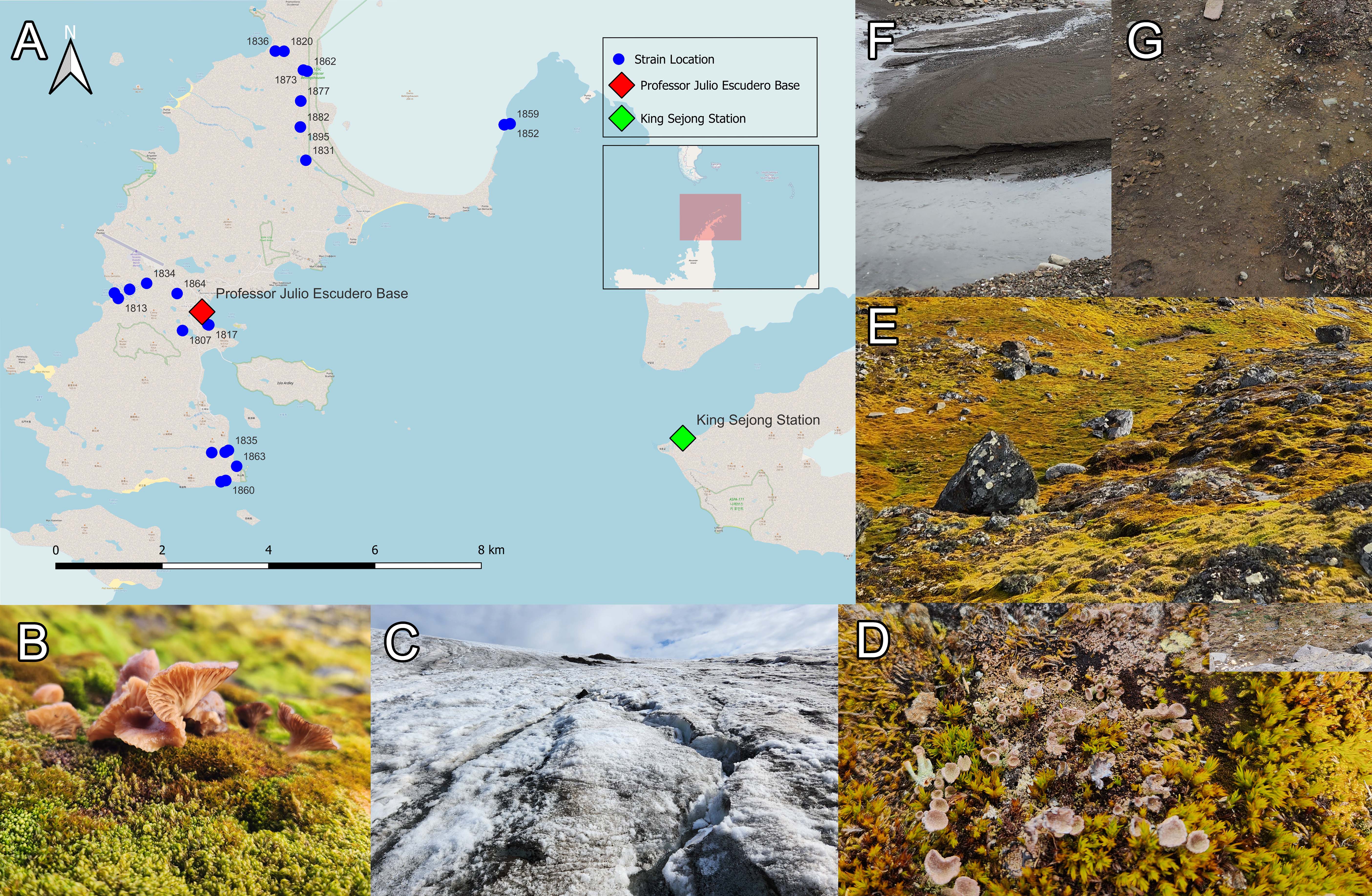
Antarctic samples for fungal isolation were collected from the Fildes Peninsula, Antarctica in March 2023 (Fig. 1). The exact locations and sample types are listed in Table 1. Samples were collected in sterilized falcon tubes (28×120 mm²) using a metal spatula sterilized with 70% alcohol, transported to Julio Escudero Base Laboratories, and stored at 4°C. They were then transported to the Laboratory of Applied and Sustainable Chemistry (LabQAS; Universidad del Bío-Bío, Chile).
A measured amount (5 g) of each collected sample (sediment, soil, moss, and fruiting body) was resuspended in 10 ml of sterile Type I ultrapure water. From the resulting suspension, 500 µl was plated on Potato Dextrose Agar (PDA; Difco, USA) supplemented with 100 mg/ml tetracycline and 100 mg/ml streptomycin to prevent bacterial contamination and incubated at 13–17°C for one week. Endophytic fungi were isolated from Deschampsia antarctica followed the method outlined by Ismail et al. (2021), with some modifications. Briefly, approximately 5 g of root was washed under running tap water to remove any residual soil. Roots that died or showed signs of lesions or discoloration were excluded from the study. The remaining healthy roots were surface sterilized by immersion in 70% ethanol for 3 min, followed by a 2.5 min soak in sodium hypochlorite solution (approximately 5% active chlorine). The roots were then rinsed three times with sterile Type I ultrapure water for 3 min. After surface sterilization, the roots were dried on sterile filter paper and cut into small segments. Seventy root segments per plot were placed on PDA media supplemented with 100 µg/ml tetracycline and 100 mg/ml streptomycin to inhibit bacterial growth. The plates were incubated at 15°C for 4 weeks. The cultures were carefully monitored for fungal mycelia emergence. Once the mycelia were observed, they were immediately transferred to fresh PDA plates to encourage further growth.
Distinct fungal colonies were selected based on their morphological characteristics, including colony color, texture, border type, and radial growth rate. These distinct colonies were then sub-cultured on fresh PDA plates to obtain pure fungal strains. All fungal strains were deposited in the LabQAS Fungal Collection at the Universidad del Bío-Bío, Chile.
Molecular identification
Genomic DNA was extracted from lyophilized tissues of each fungal strain grown on PDA (using 5 mm diameter blocks) using an AccuPrep Genomic DNA extraction kit (Bioneer Co., Korea), following the manufacturer’s instructions, with a modification of the CTAB buffer instead of the TL buffer. Polymerase chain reaction (PCR) was performed on a C1000 thermal cycler (Bio-Rad, USA) using the AccuPower PCR premix (Bioneer Co., Korea). The primer sets ITS1 and ITS4 (White et al., 1990) were used to amplify the ITS region for all fungal strains under the following conditions: 95°C for 5 min; 35 cycles of 95°C for 40 s, 55°C for 40 s, and 72°C for 1 min; and 72°C for 5 min. All PCR products were verified by gel electrophoresis on a 1% agarose gel and Gel Doc™ XR (Bio-Rad, USA). The PCR products were purified using the Expin™ PCR Purification Kit (GeneAll Biotechnology Co., Korea). DNA sequencing was performed with the same primers used for PCR by Macrogen (Korea), using an ABI PRISM 3700 Genetic Analyzer (Life Technologies, USA). The resulting sequences were proofread and manually edited using Geneious Prime software ver. 2024.0.7 (Biomatters Ltd., USA; Kearse et al., 2012). The forward and reverse sequences obtained were assembled using the de novo assembly function in Geneious Prime software ver. 2024.0.7 (Biomatters Ltd., USA; Kearse et al., 2012).
Preliminary identification at a higher taxonomic level (mostly at the genus level; if not possible, then at the family level) was performed using NCBI BLAST with the ITS region sequences. Based on the preliminary identification via NCBI BLAST, appropriate additional genetic markers for each genus were selected through a reference search to allow for species-level identification (Table S1). The PCR conditions for each primer set are summarized in Table S1. The generated sequences were sequenced and edited according to the same protocol used to generate the ITS sequences. All newly generated sequences were deposited in GenBank (Table 1).
For phylogeny-based identification, reference sequences (mostly holotype sequences) were retrieved from GenBank. When holotype sequences were unavailable, verified strain sequences from the published literature were used (Table S2). Using both reference sequences and the newly generated sequences, phylogenetic analyses were performed using FunVIP 0.3.19 with the ‘--preset fast’ setting, employing FastTree for tree construction (https://github.com/Changwanseo/FunVIP; Seo et al., under Review). The set of genetic markers used for the final identification varied depending on the genus. The final species assignment was validated based on phylogenetic evidence, specifically the branch length and local support values of the phylogenetic tree generated using FastTree v.2.1.11 (Price et al., 2010).
To construct the phylogenetic tree shown in Fig. 2, RAxML phylogenetic analysis was conducted using the GTR+GAMMA model with 1,000 replicates using RAxML ver. 8 (Stamatakis, 2014). The analysis incorporated the ITS and LSU sequences of the strains obtained in this study, along with two outgroup sequences, Conidiobolus coronatus AFTOL-ID 137 and Entomophaga maimaiga ARSEF 1400 (Gryganskyi et al., 2012).
Antibiosis assay employing dual-culture method against B. cinerea and F. culmorum
Antarctic fungal strains were evaluated against two pathogenic fungi, B. cinerea and F. culmorum. For in vitro assays, the strain of B. cinerea F003 was obtained in 2006 from the blueberry fruit cv. O’Neal, infected with this fungus, in Chillán, Ñuble Region, Chile. The strain was identified based on its microscopic morphological characteristics (presence of conidia and conidiophores) and confirmed by PCR using specific primers Bc3F/R, which amplify the intergenic spacer (IGS) region of the ribosomal DNA of B. cinerea (Suarez et al., 2005). The pathogenic isolate of F. culmorum strain F066 was isolated from European hazelnut cv. Barcelona in Camarico, Maule Region, Chile. Identification was based on the microscopic morphological characteristics and phylogenetic analysis of the ITS (MT640271), RPB2 (MT997139), TEF1 (MT661593), and CAL (MT997140) regions (Mishra et al., 2000; O'Donnell et al., 2000, 2008).
Mycelial disks (5 mm in diameter) of Antarctic fungal strains and pathogens were obtained from the margin of an actively growing culture using a cork borer. Both mycelial disks were placed on a Petri dish with 15 ml of PDA and positioned 6 cm apart. The negative controls consisted of mycelial disks from the pathogen alone. The plates were incubated in dark in a culture chamber at 25°C. The percentage of inhibition of radial growth (PIRG) was calculated using the following equation:
where PIRG is the percentage of growth inhibition,
Dc is the growth (mm) of the pathogenic fungus in the control group.
Dt is the pathogen growth (mm) in the presence of an Antarctic fungus.
Three replicates were performed for each treatment group. Antagonistic activity was evaluated by measuring the growth radius of the pathogenic fungal mycelia. Once the pathogenic fungus grew free of competition (negative control) and occupied the entire plate, the experiment was terminated. Fusarium culmorum and B. cinerea occupied the entire plate in 15 and 10 days, respectively.
Results
Identification of Antarctic fungi
A total of 97 Antarctic fungal strains were isolated from biotic (moss, lichen, fruit body, macroalgae, and root) and abiotic substrates (soil, sediment, ice, and styrofoam) in similar proportions (Fig. 2), with 48% and 40% of each substrate type, respectively. The substrate type with the highest number of fungal strains was soil (20 strains), followed by moss (18 strains), and roots (16 strains; Table 1).
The ITS region sequences were successfully obtained from 95 of the 97 strains. NCBI BLAST analysis was performed using the ITS region of these 95 strains, whereas the LSU region was used for the remaining two strains. This preliminary analysis identified 97 strains representing 58 taxa. Among these, 54 taxa were assigned to 19 known genera, whereas the remaining four taxa could not be assigned to any known genera. These four taxa matched annotated fungal sequences in the NCBI BLAST database: “Dothideomycetes sp.” (strain numbers: 1808, 1816), “Fungal sp.” (1818, 1824), “Helotiales sp.” (1812, 1813, 1830, 1855, 1859, 1884), and “Uncultured endophytic fungi” (1822, 1842).
Based on previous studies, additional genetic markers suitable for each taxonomic classification were selected, and 132 additional genetic marker sequences were acquired (Table 1): 44 sequences in the LSU region, 34 in the TUB region, 7 in the CMD region, 10 in the ACT region, 34 in the TEF1 region, and 3 in the RPB2 region. Phylogenetic analysis using multiple genetic markers was conducted, along with the appropriate reference sequences for each genus. The analysis confirmed that the 58 taxa belonged to three phyla: 6 classes (2 isolates in Agaricomycetes, 8 in Dothideomycetes, 22 in Eurotiomycetes, 47 in Leotiomycetes, 3 in Mortierellomycetes, and 15 in Sordariomycetes), 12 orders (1 isolate in Agaricales, 1 in Amphisphaeriales, 1 in Atheliales, 5 in Cladosporiales, 21 in Eurotiales, 21 in Helotiales, 14 in Hypocreales, 3 in Mortierellales, 1 in Onygenales, 3 in Pleosporales, and 26 in Thelebolales), 21 families, and 23 genera (Fig. S1). Approximately 30% of the 97 strains (29 strains) were identified at the species-level, whereas the remaining 70% were confirmed as new species candidates, particularly those concentrated in Leotiomycetes. The complete strain phylogeny is presented in Fig. 2, based on ITS and LSU sequences, and the final identification results are reflected in the strain annotations.
Antibiosis evaluation of Antarctic fungi against B. cinerea and F. culmorum
Using a dual-culture assay approach and PIRG as a quantifiable variable, we evaluated the antibiosis potential of all isolated fungal strains against B. cinerea and F. culmorum (Tables S3 and S4). Overall, remarkable antibiosis bioactivities were observed in the isolated fungal strains, with the best examples shown in Fig. 3.
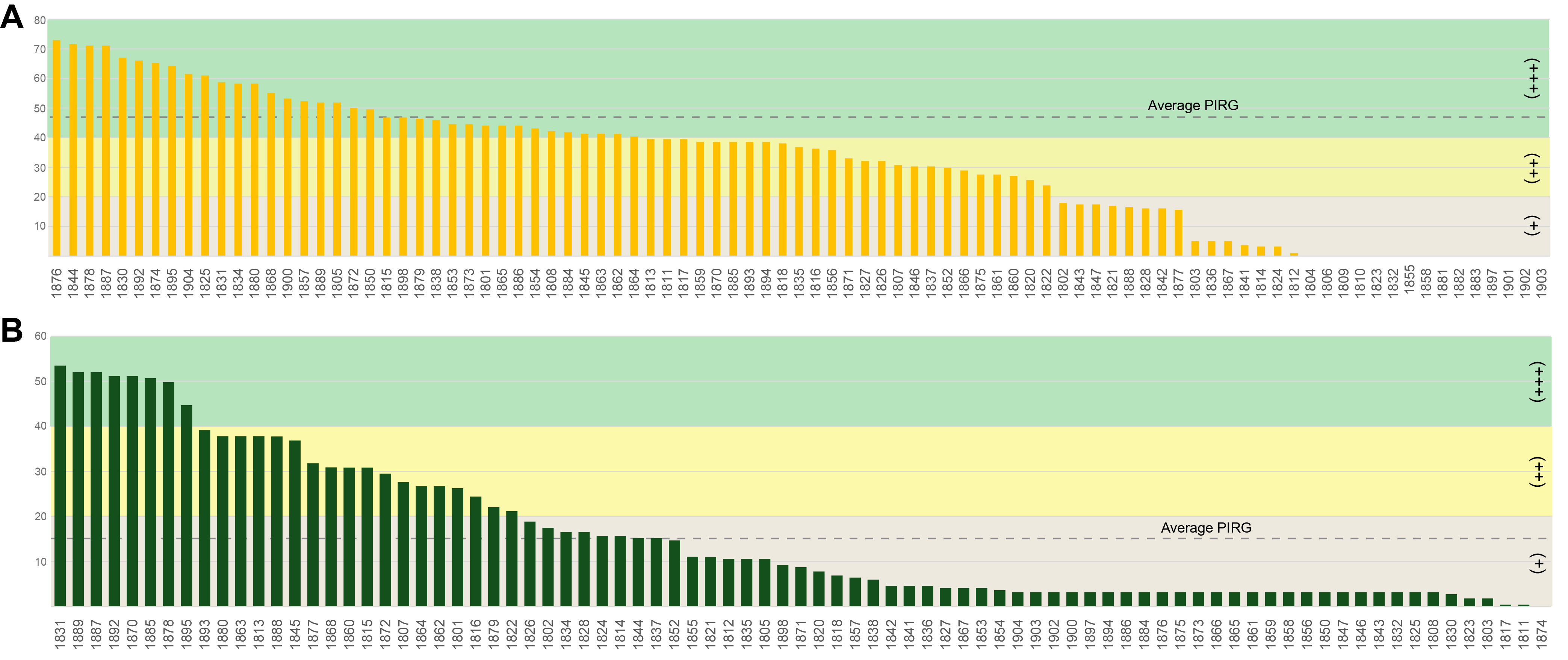
The isolated Antarctic fungi exhibited antibiosis activity against B. cinerea and F. culmorum, with PIRG values ranging from 0 to 72.95% and from 0 to 53.45%, respectively (Tables S3 and S4). Based on the PIRG values, antibiosis activity was categorized into four levels: +++ (PIRG: >40%), ++ (PIRG: 20–40%), + (PIRG: 0–20%), and 0 (no inhibition). The strains showing the highest level of inhibition (+++) included 36 and 8 strains against B. cinerea and F. culmorum, respectively (Tables S3 and S4). The antibiosis activity of the isolated Antarctic fungi was, on average, higher against B. cinerea than against F. culmorum (Fig. 4). However, the antibiosis activity of each fungal strain against the two plant pathogenic fungi did not always align consistently.
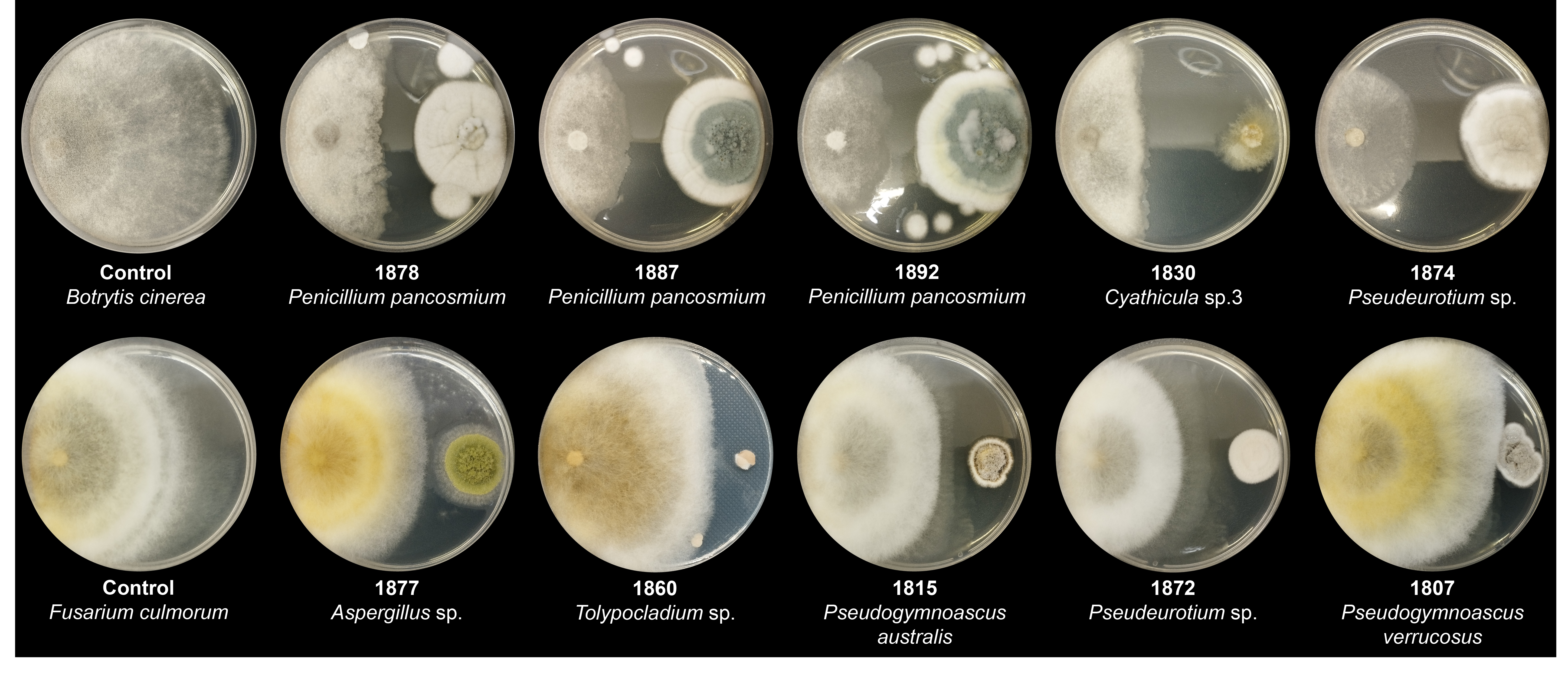
Three Penicillium pancosmium strains, 1878, 1887, and 1892, showed elevated levels of antibiosis against B. cinerea, with PIRG values of 71.1, 71.1, and 66, respectively. Additionally, new species candidates of the genera Cyathicula (1830) and Pseudeurotium (1874) showed remarkable levels of antibiosis activity, with PIRG values of 66.9 and 65.1, respectively (Fig. 4A, Table S3). Against F. culmorum, the new species candidates of Aspergillus (1877) and Tolypocladium (1860) most actively controlled pathogen growth, with PIRG values of 31.8 and 30.8, respectively. In addition, two Pseudogymnoascus species (1815 and 1807) and one Pseudeurotium strain (1872) controlled F. culmorum growth (PIRG = 30.8, 27.6, and 29.5, respectively; Fig. 4B, Table S4).
Discussion
We successfully isolated 58 diverse fungal taxa at the species level from various regions and substrates in Antarctica, representing the first report of culturable fungi associated with Antarctic fruiting bodies. Additionally, we evaluated the antibiosis potential of all fungal strains isolated during the Antarctic expedition (ECA 59) against B. cinerea and F. culmorum. This study revealed several Antarctic strains that substantially inhibited the growth of agriculturally relevant fungal pathogens, thereby emphasizing their ecological and biotechnological significance.
A significant number of these isolated fungal strains were identified as new species candidates because they showed no match at the species-level in the existing species databases. This highlights the lack of comprehensive taxonomic studies on Antarctic fungi and their underrepresentation in global databases. Furthermore, discrepancies between the final phylogenetic identification and ITS-based BLAST results were observed, particularly within the orders Pleosporales and Helotiales. For instance, strains preliminarily identified as “Fungal sp.” (1818 and 1824) and “Helotiales sp.” (1812, 1830, and 1859), based on ITS-based BLAST, were later classified as Cyathicula through phylogenetic analysis. A detailed taxonomic study revealed that the closest known species, Cyathicula microspora, shared only 86.1% to 92.3% ITS sequence identity with these Antarctic fungal strains, indicating a substantial genetic divergence. These findings further highlight the limitations of fungal sequence curation in the NCBI database, particularly for the identification of Antarctic fungi, due to the lack of taxonomic studies on these organisms.
Furthermore, the limitations of ITS as the sole marker and the necessity of multi-genetic approaches for accurate fungal taxonomy were also pointed out, when studying for Antarctic fungi. To overcome the taxonomic ambiguities of Antarctic fungi, we applied a multigene marker-based approach to discover new species candidates. This approach is particularly effective for genera such as Penicillium and Cladosporium, which require additional markers, such as TUB and RPB2, for reliable species-level classification (Bensch et al., 2012; Visagie et al., 2014). By applying this approach, we resolved taxonomic ambiguities and demonstrated its utility in revealing previously uncharacterized fungal diversity. By providing accurate information on these poorly studied Antarctic fungi, this study contributes to the understanding of their potential impact on the changing Antarctic ecosystem and their hidden capabilities for various future applications.
Taxonomic ambiguities in identifying Antarctic fungi were particularly pronounced in the class Leotiomycetes, a group frequently reported in polar environments, including soil, moss, and marine habitats, such as algae, seawater, and sponges (Kochkina et al., 2014, 2019; Ordóñez-Enireb et al., 2022; Rämä et al., 2017; Rosa et al., 2019, 2020). Despite their ecological significance (Bates et al., 2012; Câmara et al., 2021; Kochkina et al., 2014; Park et al., 2015; Yu et al., 2018), Leotiomycetes remain understudied, with many unresolved taxonomic issues (Johnston et al., 2019; Quandt and Haelewaters, 2021). This makes the species-level identification particularly difficult for Antarctic Leotiomycetes (Henríquez et al., 2014; Hirose et al., 2016, 2017; Kochkina et al., 2014; Ordóñez-Enireb et al., 2022). Recent studies have reported an increasing association between Antarctic mosses and Antarctic Leotiomycetes species (De Carvalho et al., 2019; Hirose et al., 2016, 2017), with some Leotiomycetes species identified as pathogenic (Rosa et al., 2020, 2021). These findings underscore the need for accurate identification within this class.
Our findings highlight the antifungal potential of Antarctic fungi, many of which are poorly understood. A dual-culture assay revealed significant antifungal activity against two major phytopathogens, B. cinerea and F. culmorum. On average, B. cinerea was more susceptible to the antibiosis effects of the Antarctic fungal isolates than F. culmorum (Fig. 4). Fungi belonging to Eurotiales, including Penicillium pancosmium, exhibit particularly strong antibiosis activity, suggesting their potential as natural fungicides. Although Penicillium species are well-documented for their biocontrol activities (Roca-Couso et al., 2021; Thambugala et al., 2020), studies on P. pancosmium remain limited, making this a notable discovery.
Among the new species candidates, strains from Cyathicula and Pseudeurotium showed the highest levels of antibiosis activity against both plant pathogens. To the best of our knowledge, this is the first report of the antifungal activity of Cyathicula. Although other species of the family Helotiaceae, to which Cyathicula belongs, also produce various secondary metabolites with antifungal properties (Chen et al., 2013; Elhamouly et al., 2022), the discovery of such activity in Cyathicula expands our understanding of the functional diversity within Helotiaceae, highlighting its potential as a source of novel antifungal compounds. Moreover, Antarctic strains of Aspergillus, Penicillium, Pseudeurotium, and Tolypocladium exhibited antibiosis activity. These fungal groups were well-known for synthesizing antifungal secondary metabolites (Bladt et al., 2013; Brown et al., 1976; Bushley et al., 2013; Heo et al., 2019; Khokhar et al., 2011; Quandt et al., 2015; Wang et al., 2023).
Notably, Pseudogymnoascus, the most taxonomically diverse genus identified in this study (6 taxa, 18 isolates), demonstrated significant antifungal activity, with most showing above-average activity against at least one plant pathogen. This aligns with the results of previous studies indicating the capacity of Pseudogymnoascus to synthesize diverse antifungal compounds, such as amphiols, geomycins A–C, and various sesquiterpenes (Antipova et al., 2023; Shi et al., 2021). These findings emphasize their potential as key sources of bioactive compounds and their ecological role in Antarctic environments, where antifungal properties may confer adaptive advantages. This study highlights the immense microbial diversity within Antarctic ecosystems and their potential to be broadly applicable in biotechnology, agriculture, and medicine. The extreme conditions in Antarctica likely drive unique selective pressures, fostering the evolution of microorganisms producing distinctive secondary metabolites (Marx et al., 2007; Núñez-Montero and Barrientos, 2018; Ramasamy et al., 2023).
Therefore, this study improves our understanding of Antarctic fungi by elucidating their diversity across various Antarctic habitats and their antibiosis activity against plant pathogenic fungi. Furthermore, this study highlights the importance of applying multi-genetic approaches for the accurate identification and taxonomic classification of fungi in underexplored regions, such as Antarctica. By identifying new species candidates and characterizing their antibiosis activity against B. cinerea and F. culmorum, we demonstrate the immense potential of Antarctic fungi as a source of novel bioactive compounds with profound biotechnological applications. As the Antarctic ecosystem continues to undergo changes, this study establishes a foundation for future ecological and biotechnological research by providing critical insights into fungal taxonomy and physiology.
Acknowledgments
This study was supported by the Instituto Antártico Chileno (INACH) (grant number: INACH RT_16-21), VRIP-Universidad del Bío-Bío (grant numbers: GI2310643 and EQ2326450), and the Korea Institute of Marine Science & Technology Promotion (KIMST) funded by the Ministry of Oceans and Fisheries (grant number: RS-2024-00341152).
Conflict of interest
None.
Ethical Statements
Not applicable.
Supplementary Information
The online version contains supplementary material available at https://doi.org/10.71150/jm.2411029.
Fig. S1.
Phylogenetic trees of the 97 fungal strains classified by each genus. Each tree, from A to W, is organized by genus, with the genus name, type of tree constructed, and the genetic markers used for the phylogenetic analysis indicated. Our strains are highlighted in bold, and the final identification results are marked with boxes.
jm-2411029-Supplementary-Fig-S1.pdf
Fig. 1.Sampling sites in Antarctica (ECA59 Expedition) with photographs of each sample type. (A) Map indicating the sampling sites with the location of research stations from Chile and South Korea. Representative photographs of (B) fruit body, (C) ice, (D) lichen, (E) moss, (F) sediment, and (G) soil samples.
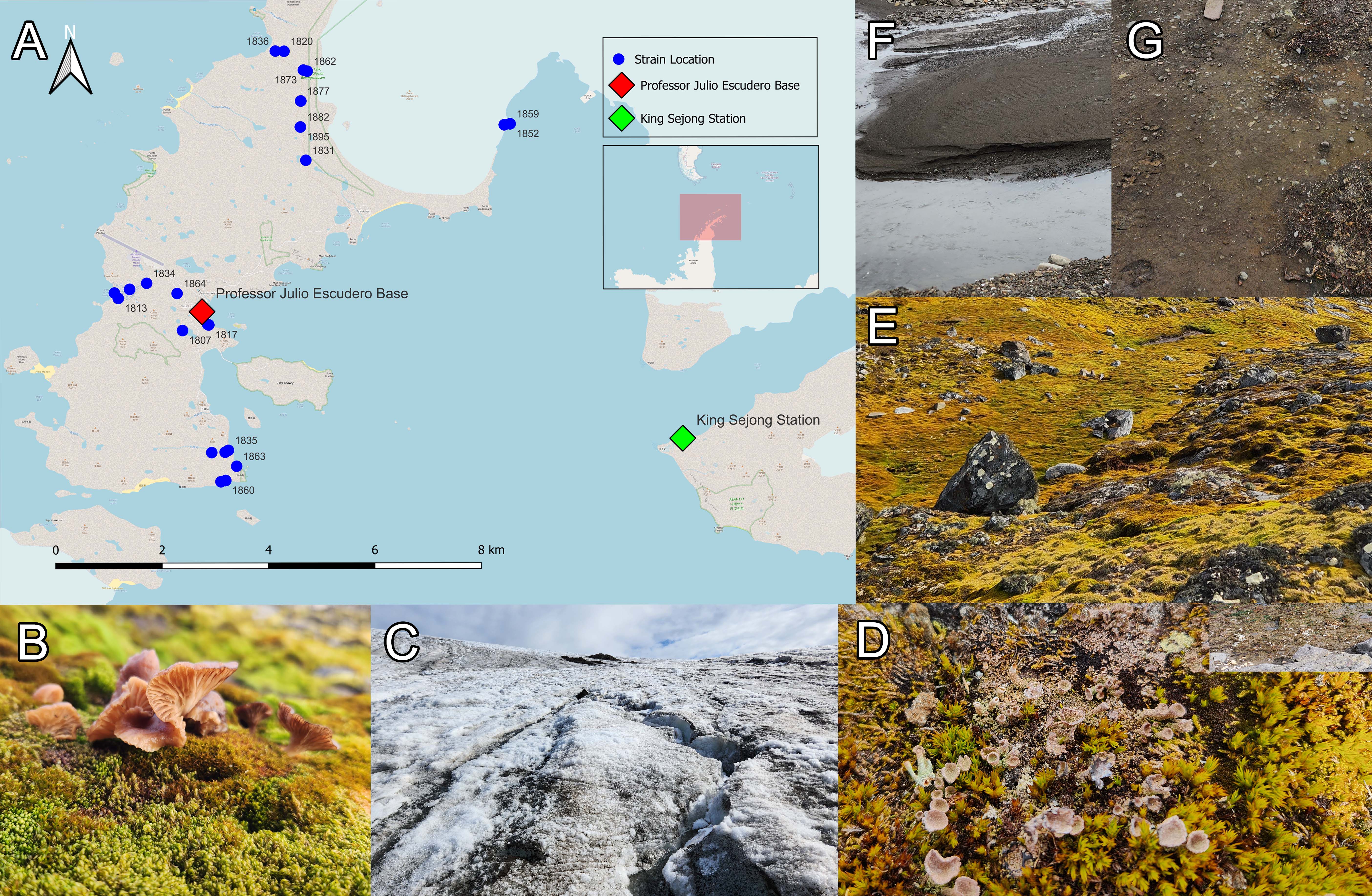
Fig. 2.Phylogenetic tree of 97 fungal strains isolated in this study. The phylogenetic tree was constructed using RAxML analysis with internal transcribed spacer (ITS) and LSU sequences. The final identification results for each strain are shown along with the strain numbers in bold. Bootstrap values greater than 70% are indicated at each branch node, and branches with a bootstrap value of 100 are represented by thick lines. The substrate type from which each strain was isolated is indicated next to the strain, with the corresponding type highlighted by a colored box. For clarity, the substrate types are listed at the top of each column.
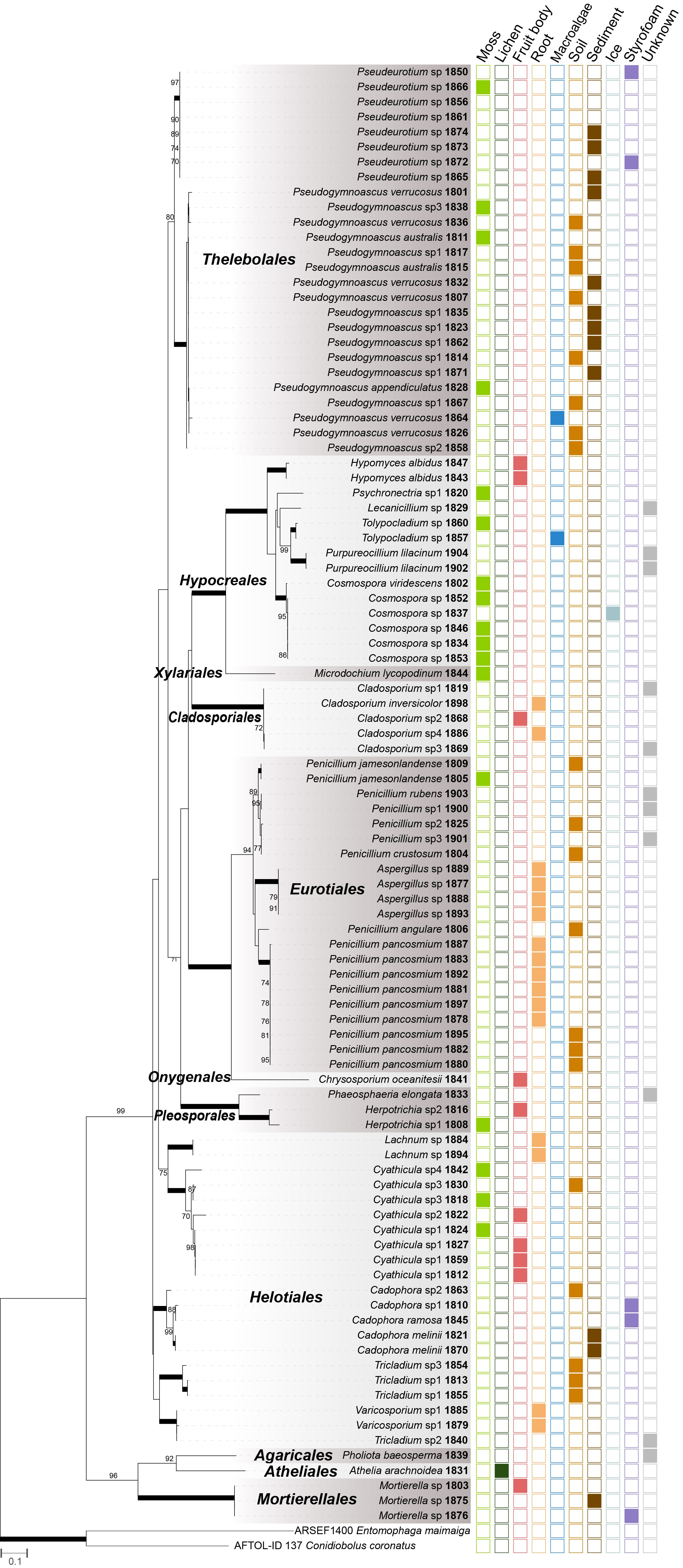
Fig. 3.Images showing the top five strains with the highest antibiosis activity against two plant pathogens. The leftmost image in each row represents the control for B. cinerea and F. culmorum. Strain numbers and identification results are indicated below each plate.
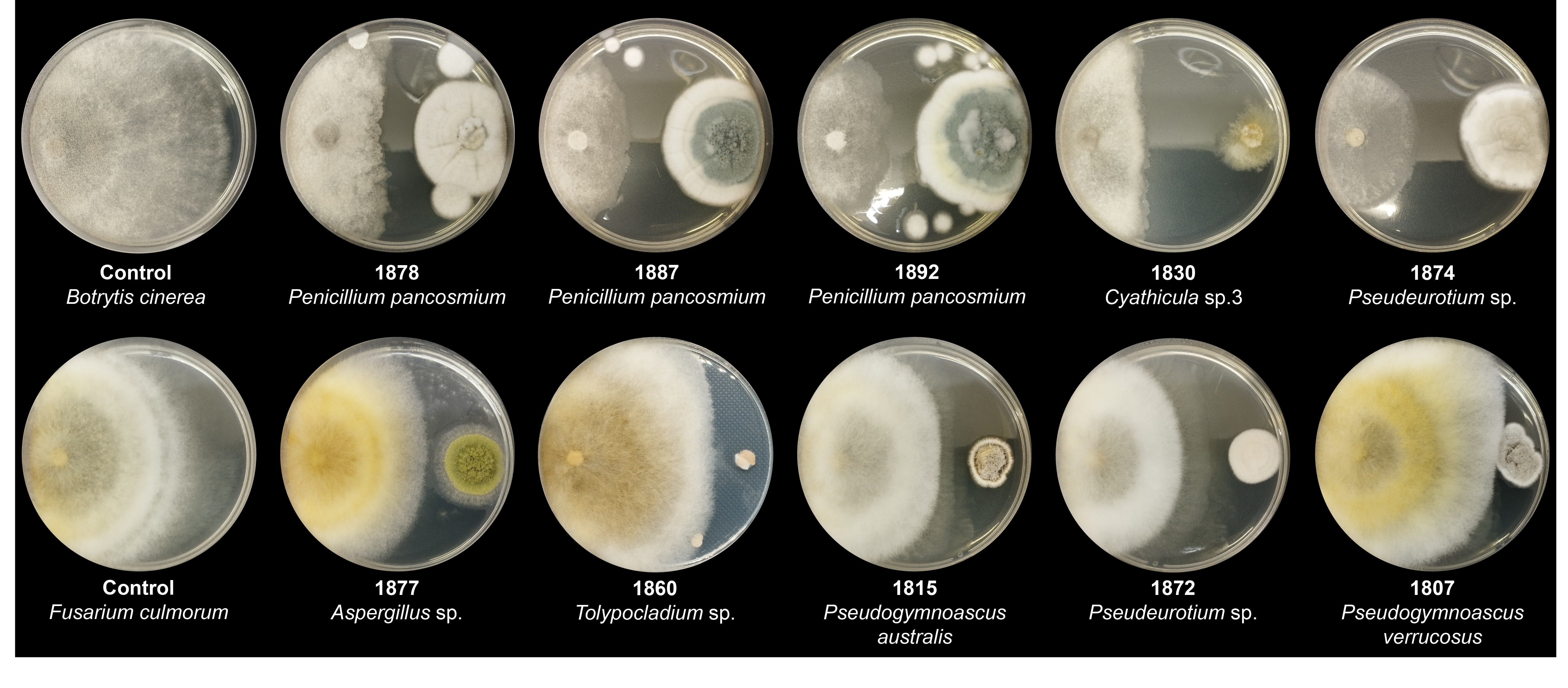
Fig. 4.Antibiosis activity of Antarctic fungal strains against two plant pathogens. The graphs show the antibiosis activity results, ranked from highest to lowest, for (A) B. cinerea and (B) F. culmorum. The average percentage of inhibition of radial growth (PIRG) is indicated by the dashed line in each graph. Antibiosis activity is categorized into four levels, represented by distinct colors: (+++) in green, (++) in yellow, (+) in light gray, and below-average in gray.
Table 1.Collection information and accession numbers of the 97 fungal strains isolated in this study
|
Order |
Family |
Species identification |
Strain NUM |
Substrate |
Latitude (S) |
Longitude (W) |
ITS |
LSU |
TUB
|
CMD
|
ACT
|
TEF1
|
RPB2
|
|
Agaricales |
Strophariaceae |
Pholiota baeosperma
|
1839 |
- |
- |
- |
PQ427716 |
PQ427669 |
|
|
|
|
|
|
Amphisphaeriales |
Amphisphaeriaceae |
Microdochium lycopodinum
|
1844 |
Moss |
62°10'11.4168" |
58°51'10.2096" |
PQ427765 |
PQ427672 |
|
|
|
|
|
|
Atheliales |
Atheliaceae |
Athelia arachnoidea
|
1831 |
Lichen |
62°10'32.9340" |
58°55'28.6860" |
PQ427721 |
|
|
|
|
|
|
|
Cladosporiales |
Cladosporiaceae |
Cladosporium inversicolor
|
1898 |
Root |
62°09'56.8980" |
58°55'35.2488" |
PQ427783 |
|
|
|
|
PQ433547 |
|
|
|
Cladosporium sp.1 |
1819 |
- |
- |
- |
PQ427699 |
|
|
|
|
|
|
|
|
Cladosporium sp.2 |
1868 |
Fruit body |
62°10'10.8408" |
58°51'02.6460" |
PQ427748 |
|
|
|
PQ433531 |
PQ433545 |
|
|
|
Cladosporium sp.3 |
1869 |
- |
- |
- |
PQ427747 |
|
|
|
|
|
|
|
|
Cladosporium sp.4 |
1886 |
Root |
62°09'56.8980" |
58°55'35.2488" |
PQ427692 |
|
|
|
PQ433530 |
PQ433546 |
|
|
Eurotiales |
Aspergillaceae |
Aspergillus sp. |
1877 |
Root |
62°09'56.8980" |
58°55'35.2488" |
PQ427757 |
|
PQ456765 |
|
|
|
|
|
|
|
1888 |
Root |
62°09'56.8980" |
58°55'35.2488" |
PQ427709 |
|
|
|
|
|
|
|
|
|
1889 |
Root |
62°09'56.8980" |
58°55'35.2488" |
PQ427694 |
|
PQ456764 |
|
|
|
|
|
|
|
1893 |
Root |
62°09'56.8980" |
58°55'35.2488" |
PQ427710 |
|
|
|
|
|
|
|
|
Penicillium angulare
|
1806 |
Soil |
62°10'11.4168" |
58°51'10.2096" |
PQ427704 |
|
PQ456772 |
|
|
|
|
|
|
Penicillium crustosum
|
1804 |
Soil |
62°13'48.2520" |
58°57'19.5336" |
PQ427740 |
|
PQ456773 |
|
|
|
|
|
|
Penicillium jamesonlandense
|
1805 |
Moss |
62°10'11.4168" |
58°51'10.2096" |
PQ427738 |
|
PQ456774 |
|
|
|
|
|
|
|
1809 |
Soil |
62°10'11.4168" |
58°51'10.2096" |
PQ427739 |
|
PQ456775 |
|
|
|
|
|
|
Penicillium pancosmium
|
1878 |
Root |
62°10'11.4168" |
58°51'10.2096" |
PQ427742 |
|
PQ456783 |
|
|
|
|
|
|
|
1880 |
Soil |
62°10'12.7" |
58°55'35.8" |
PQ427717 |
|
PQ456780 |
PQ433532 |
|
|
|
|
|
|
1881 |
Root |
62°10'12.7" |
58°55'35.8" |
PQ427707 |
|
PQ456778 |
PQ433533 |
|
|
|
|
|
|
1882 |
Soil |
62°10'12.7" |
58°55'35.8" |
PQ427697 |
|
PQ456777 |
|
|
|
|
|
|
|
1883 |
Root |
62°09'56.8980" |
58°55'35.2488" |
PQ427689 |
|
PQ456776 |
PQ433534 |
|
|
|
|
|
|
1887 |
Root |
62°09'56.8980" |
58°55'35.2488" |
PQ427737 |
|
PQ456782 |
PQ433535 |
|
|
|
|
|
|
1892 |
Root |
62°09'56.8980" |
58°55'35.2488" |
PQ427708 |
|
PQ456779 |
PQ433536 |
|
|
|
|
|
|
1895 |
Soil |
62°10'12.7" |
58°55'35.8" |
PQ427743 |
|
PQ456784 |
|
|
|
|
|
|
|
1897 |
Root |
62°09'56.8980" |
58°55'35.2488" |
PQ427736 |
|
PQ456781 |
PQ433537 |
|
|
|
|
|
Penicillium rubens
|
1903 |
- |
- |
- |
PQ427741 |
|
PQ456785 |
PQ433538 |
|
|
|
|
|
Penicillium sp.1 |
1900 |
- |
- |
- |
PQ427696 |
|
PQ456786 |
|
|
|
|
|
|
Penicillium sp.2 |
1825 |
Soil |
62°12'16.3656" |
58°58'09.4368" |
PQ427735 |
|
PQ456787 |
|
|
|
|
|
|
Penicillium sp.3 |
1901 |
- |
- |
- |
PQ427773 |
|
PQ456788 |
|
|
|
|
|
Helotiales |
Discinellaceae |
Varicosporium sp. |
1879 |
Root |
62°10'11.4168" |
58°51'10.2096" |
PQ427701 |
PQ427685 |
|
|
|
|
|
|
|
|
1885 |
Root |
62°10'11.4168" |
58°51'10.2096" |
PQ427763 |
PQ427686 |
|
|
|
|
|
|
Helotiaceae |
Cyathicula sp.1 |
1812 |
Fruit body |
62°13'47.4816" |
58°57'13.3560" |
PQ427754 |
|
|
|
|
|
|
|
|
|
1824 |
Moss |
62°11'51.3420" |
58°59'18.3408" |
PQ427753 |
|
|
|
|
|
|
|
|
|
1827 |
Fruit body |
62°13'47.4816" |
58°57'13.3560" |
PQ427732 |
|
|
|
|
|
|
|
|
|
1859 |
Fruit body |
62°10'10.8408" |
58°51'02.6460" |
PQ427777 |
|
|
|
|
|
|
|
|
Cyathicula sp.2 |
1822 |
Fruit body |
62°12'12.69" |
58°57'36.59" |
PQ427760 |
|
|
|
|
|
|
|
|
Cyathicula sp.3 |
1818 |
Moss |
62°12'12.9708" |
58°57'35.2908" |
PQ427713 |
|
|
|
|
|
|
|
|
|
1830 |
Soil |
62°12'12.9708" |
58°57'35.2908" |
PQ427734 |
|
|
|
|
|
|
|
|
Cyathicula sp.4 |
1842 |
Moss |
62°09'26.6148" |
58°55'57.0360" |
PQ427755 |
|
|
|
|
|
|
|
Lachnaceae |
Lachnum sp. |
1884 |
Root |
62°09'56.8980" |
58°55'35.2488" |
PQ427781 |
|
|
|
|
|
|
|
|
|
1894 |
Root |
62°10'11.4168" |
58°51'10.2096" |
PQ427782 |
|
|
|
|
|
|
|
Ploettnerulaceae |
Cadophora melinii
|
1821 |
Sediment |
62°11'51.3420" |
58°59'18.3408" |
PQ427691 |
PQ427659 |
PQ433548 |
|
|
|
|
|
|
|
1870 |
Sediment |
62°11'51.3420" |
58°59'18.3408" |
PQ427759 |
|
PQ433549 |
|
|
|
|
|
|
Cadophora ramosa
|
1845 |
Styrofoam |
- |
- |
PQ427705 |
PQ427673 |
PQ433550 |
|
|
|
|
|
|
Cadophora sp.1 |
1810 |
Styrofoam |
- |
- |
PQ427756 |
|
|
|
|
|
|
|
|
Cadophora sp.2 |
1863 |
Soil |
62°13'38.8308" |
58°56'59.0640" |
PQ427690 |
PQ427679 |
|
|
|
|
|
|
Tricladiaceae |
Tricladium sp.1 |
1813 |
Soil |
62°11'56.83" |
58°59'33.12" |
PQ427714 |
|
|
|
|
|
|
|
|
Tricladium sp.1 |
1855 |
Soil |
62°11'56.83" |
58°59'33.12" |
PQ427719 |
|
|
|
|
|
|
|
|
Tricladium sp.2 |
1840 |
- |
- |
- |
|
PQ427670 |
|
|
|
|
|
|
|
Tricladium sp.3 |
1854 |
Soil |
62°12'12.9708" |
58°57'35.2908" |
PQ427776 |
PQ427675 |
|
|
|
|
|
|
Hypocreales |
Cordycipitaceae |
Lecanicillium sp. |
1829 |
- |
- |
- |
PQ427751 |
PQ427663 |
|
|
|
|
|
|
Hypocreaceae |
Hypomyces albidus
|
1843 |
Fruit body |
62°10'10.8408" |
58°51'02.6460" |
PQ427770 |
|
|
|
|
PQ433543 |
PQ433539 |
|
|
|
1847 |
Fruit body |
62°10'10.8408" |
58°51'02.6460" |
PQ427766 |
|
|
|
|
PQ433544 |
PQ433540 |
|
Nectriaceae |
Cosmospora viridescens
|
1802 |
Moss |
62°13'47.4816" |
58°57'13.3560" |
PQ427764 |
|
PQ456771 |
|
|
|
|
|
|
Cosmospora sp. |
1834 |
Moss |
62°11'47.6700" |
58°58'56.0928" |
PQ427768 |
|
PQ456770 |
|
|
PQ433542 |
|
|
|
|
1837 |
Ice |
62°13'30.5256" |
58°57'31.5036" |
PQ427767 |
|
PQ456769 |
|
|
|
|
|
|
|
1846 |
Moss |
62°13'47.4816" |
58°57'13.3560" |
PQ427695 |
|
PQ456766 |
|
|
|
|
|
|
|
1852 |
Moss |
62°10'10.8408" |
58°51'02.6460" |
PQ427726 |
|
PQ456768 |
|
|
|
|
|
|
|
1853 |
Moss |
62°11'51.3420" |
58°59'18.3408" |
PQ427712 |
|
PQ456767 |
|
|
|
|
|
Tilachlidiaceae |
Psychronectria sp. |
1820 |
Moss |
62°09'26.6148" |
58°55'57.0360" |
PQ427700 |
|
|
|
|
|
|
|
Ophiocordycipitaceae |
Purpureocillium lilacinum
|
1902 |
- |
- |
- |
PQ427702 |
PQ427687 |
|
|
|
|
|
|
|
|
1904 |
- |
- |
- |
PQ427693 |
PQ427688 |
|
|
|
|
|
|
|
Tolypocladium sp. |
1857 |
Macroalga |
62°10'10.8408" |
58°51'02.6460" |
PQ427703 |
PQ427676 |
|
|
|
|
|
|
|
|
1860 |
Moss |
62°13'47.4816" |
58°57'13.3560" |
PQ427728 |
|
|
|
|
|
|
|
Mortierellales |
Mortierellaceae |
Mortierella sp. |
1803 |
Fruit body |
62°13'47.4816" |
58°57'13.3560" |
PQ427746 |
|
|
|
|
|
|
|
|
|
1875 |
Lagoon sediment |
62°12'16.3656" |
58°58'09.4368" |
PQ427722 |
|
|
|
|
|
|
|
|
|
1876 |
Styrofoam |
- |
- |
PQ427769 |
PQ427684 |
|
|
|
|
|
|
Onygenales |
Onygenaceae |
Chrysosporium sp. |
1841 |
Fruit body |
62°13'47.4816" |
58°57'13.3560" |
PQ427698 |
PQ427671 |
|
|
|
|
|
|
Pleosporales |
Melanommataceae |
Herpotrichia sp.1 |
1808 |
Moss |
62°13'47.4816" |
58°57'13.3560" |
PQ427761 |
PQ427653 |
|
|
|
PQ433541 |
|
|
|
Herpotrichia sp.2 |
1816 |
Fruit body |
62°13'47.4816" |
58°57'13.3560" |
PQ427774 |
PQ427657 |
|
|
|
|
|
|
Phaeosphaeriaceae |
Phaeosphaeria sp. |
1833 |
- |
- |
- |
|
PQ427665 |
|
|
|
|
|
|
Thelebolales |
Pseudeurotiaceae |
Pseudeurotium sp. |
1850 |
Styrofoam |
- |
- |
PQ427745 |
PQ427674 |
|
|
|
|
|
|
|
|
1856 |
Moss |
62°12'12.69" |
58°57'36.59" |
PQ427706 |
|
|
|
|
|
|
|
|
|
1861 |
Ice |
62°13'30.5256" |
58°57'31.5036" |
PQ427771 |
|
|
|
|
|
|
|
|
|
1865 |
Sediment |
62°12'12.69" |
58°57'36.59" |
PQ427779 |
|
|
|
|
|
|
|
|
|
1866 |
Moss |
62°15'34.8" |
58°59'11.16" |
PQ427778 |
PQ427681 |
|
|
|
|
|
|
|
|
1872 |
Styrofoam |
62°12'16.3656" |
58°58'09.4368" |
PQ427780 |
|
|
|
|
|
|
|
|
|
1873 |
Sediment |
62°09'38.0520" |
58°55'31.8540" |
PQ427772 |
|
|
|
|
|
|
|
|
|
1874 |
Sediment |
62°15'34.62" |
58°59'10.56" |
PQ427744 |
|
|
|
|
|
|
|
|
Pseudogymnoascus appendiculatus
|
1828 |
Moss |
62°11'51.3420" |
58°59'18.3408" |
PQ427731 |
PQ427662 |
|
|
|
|
|
|
|
Pseudogymnoascus australis
|
1811 |
Moss |
62°13'47.4816" |
58°57'13.3560" |
PQ427729 |
PQ427654 |
|
|
|
|
|
|
|
|
1815 |
Soil |
62°10'11.4168" |
58°51'10.2096" |
PQ427727 |
PQ427656 |
|
|
|
|
|
|
|
Pseudogymnoascus verrucosus
|
1801 |
Sediment |
62°11'53.5776" |
58°59'38.1156" |
PQ427752 |
PQ427651 |
|
|
|
|
|
|
|
|
1807 |
Soil |
62°12'16.3656" |
58°58'09.4368" |
PQ427730 |
PQ427652 |
|
|
|
|
|
|
|
|
1826 |
Soil |
62°12'12.9708" |
58°57'35.2908" |
PQ427723 |
PQ427661 |
|
|
|
|
|
|
|
|
1832 |
River sediment |
62°13'30.2916" |
58°57'14.0508" |
PQ427725 |
PQ427664 |
|
|
|
|
|
|
|
|
1836 |
Soil |
62°09'26.4996" |
58°56'08.3868" |
PQ427758 |
PQ427667 |
|
|
|
|
|
|
|
|
1864 |
Green alga |
62°11'53.8908" |
58°58'16.4640" |
PQ427750 |
PQ427680 |
|
|
|
|
|
|
|
Pseudogymnoascus sp.1 |
1814 |
Soil |
62°13'48.2520" |
58°57'19.5336" |
PQ427711 |
PQ427655 |
|
|
|
|
|
|
|
|
1817 |
Soil |
62°12'12.9708" |
58°57'35.2908" |
PQ427720 |
PQ427658 |
|
|
|
|
|
|
|
|
1823 |
Sediment |
62°11'53.5776" |
58°59'38.1156" |
PQ427724 |
PQ427660 |
|
|
|
|
|
|
|
|
1835 |
River sediment |
62°13'29.1360" |
58°57'09.7236" |
PQ427733 |
PQ427666 |
|
|
|
|
|
|
|
|
1862 |
Sediment |
62°09'38.7468" |
58°55'26.8320" |
PQ427749 |
PQ427678 |
|
|
|
|
|
|
|
|
1867 |
Soil |
62°12'12.9708" |
58°57'35.2908" |
PQ427715 |
PQ427682 |
|
|
|
|
|
|
|
|
1871 |
River sediment |
62°13'30.2916" |
58°57'14.0508" |
PQ427718 |
PQ427683 |
|
|
|
|
|
|
|
Pseudogymnoascus sp.2 |
1858 |
Soil |
62°12'12.9708" |
58°57'35.2908" |
PQ427762 |
PQ427677 |
|
|
|
|
|
|
|
Pseudogymnoascus sp.3 |
1838 |
Moss |
62°11'51.3420" |
58°59'18.3408" |
PQ427775 |
PQ427668 |
|
|
|
|
|
References
- Aly AH, Debbab A, Proksch P. 2011. Fifty years of drug discovery from fungi. Fungal Divers. 50: 3–19. ArticlePDF
- Antipova TV, Zaitsev KV, Zhelifonova VP, Tarlachkov SV, Grishin YK, et al. 2023. The potential of Arctic Pseudogymnoascus fungi in the biosynthesis of natural products. Fermentation. 9(8): 702.Article
- Bar-On YM, Phillips R, Milo R. 2018. The biomass distribution on Earth. Proc Natl Acad Sci USA. 115(25): 6506–6511. ArticlePubMedPMC
- Bates ST, Berg-Lyons D, Lauber CL, Walters WA, Knight R, et al. 2012. A preliminary survey of lichen-associated eukaryotes using pyrosequencing. Lichenologist. 44(1): 137–146. Article
- Bensch K, Braun U, Groenewald JZ, Crous PW. 2012. The genus Cladosporium. Stud Mycol. 72: 1–401. ArticlePubMedPMC
- Bhattarai K, Bhattarai K, Kabir ME, Bastola R, Baral B. 2021. Fungal natural products galaxy: Biochemistry and molecular genetics toward blockbuster drugs discovery. Adv Genet. 107: 193–284. ArticlePubMed
- Bladt T, Frisvad J, Knudsen P, Larsen T. 2013. Anticancer and antifungal compounds from Aspergillus, Penicillium, and other filamentous fungi. Molecules. 18(9): 11338–11376. ArticlePubMedPMC
- Brown AG, Smale TC, King TJ, Hasenkamp R, Thompson RH. 1976. Crystal and molecular structure of compactin, a new antifungal metabolite from Penicillium brevicompactum. J Chem Soc Perkin Trans 1. 11: 1165–1170. Article
- Bushley KE, Raja R, Jaiswal P, Cumbie JS, Nonogaki M, et al. 2013. The genome of Tolypocladium inflatum: Evolution, organization, and expression of the cyclosporin biosynthetic gene cluster. PLoS Genet. 9(6): e1003496. ArticlePubMedPMC
- Câmara PEAS, Convey P, Rangel SB, Konrath M, Barreto CC, et al. 2021. The largest moss carpet transplant in Antarctica and its bryosphere cryptic biodiversity. Extremophiles. 25(4): 369–384. ArticlePubMedPDF
- Chen L, Yue Q, Zhang X, Xiang M, Wang C, et al. 2013. Genomics-driven discovery of the pneumocandin biosynthetic gene cluster in the fungus Glarea lozoyensis. BMC Genomics. 14(1): 339.ArticlePubMedPMCPDF
- De Carvalho CR, Santiago IF, Da Costa Coelho L, Câmara PEAS, Silva MC, et al. 2019. Fungi associated with plants and lichens of Antarctica, In Rosa LH. (ed.), Fungi of Antarctica, pp. 165–199, Springer.
- Dean R, Van Kan JAL, Pretorius ZA, Hammond‐Kosack KE, Di Pietro A, et al. 2012. The top 10 fungal pathogens in molecular plant pathology. Mol Plant Pathol. 13(4): 414–430. ArticlePubMedPMC
- Dupuis JR, Roe AD, Sperling FAH. 2012. Multi-locus species delimitation in closely related animals and fungi: One marker is not enough. Mol Ecol. 21(18): 4422–4436. ArticlePubMed
- Dwivedi M, Singh P, Pandey AK. 2024. Botrytis fruit rot management: What have we achieved so far? Food Microbiol. 122: 104564.ArticlePubMed
- Elhamouly NA, Hewedy OA, Zaitoon A, Miraples A, Elshorbagy OT, et al. 2022. The hidden power of secondary metabolites in plant-fungi interactions and sustainable phytoremediation. Front Plant Sci. 13: 1044896.ArticlePubMedPMC
- Gholami-Shabani M, Shams-Ghahfarokhi M, Razzaghi-Abyaneh M. 2019. Natural product synthesis by fungi: Recent trends and future prospects. In Yadav AN, Singh S, Mishra S, Gupta A. (eds.), Recent advancement in white biotechnology through fungi, pp. 195–228, Springer.
- González J, Romero-Aguilar L, Matus-Ortega G, Pablo Pardo J, Flores-Alanis A, et al. 2020. Levaduras adaptadas al frío: El tesoro biotecnológico de la Antártica. TIP Rev Esp Cienc Quím Biol. 23.
- Gryganskyi AP, Humber RA, Smith ME, Miadlikovska J, Wu S, et al. 2012. Molecular phylogeny of the Entomophthoromycota. Mol Phylogenet Evol. 65(2): 682–694. ArticlePubMed
- Hahn M. 2014. The rising threat of fungicide resistance in plant pathogenic fungi: Botrytis as a case study. J Chem Biol. 7(4): 133–141. ArticlePubMedPMCPDF
- Hassan N, Rafiq M, Hayat M, Shah AA, Hasan F. 2016. Psychrophilic and psychrotrophic fungi: A comprehensive review. Rev Environ Sci Biotechnol. 15(2): 147–172. ArticlePDF
- Henríquez M, Vergara K, Norambuena J, Beiza A, Maza F, et al. 2014. Diversity of cultivable fungi associated with Antarctic marine sponges and screening for their antimicrobial, antitumoral and antioxidant potential. World J Microbiol Biotechnol. 30(1): 65–76. ArticlePubMedPDF
- Heo YM, Lee H, Kim K, Kwon SL, Park MY, et al. 2019. Fungal diversity in intertidal mudflats and abandoned solar salterns as a source for biological resources. Mar Drugs. 17(11): 601.ArticlePubMedPMC
- Herrera-Défaz M, Fuentealba D, Dibona-Villanueva L, Schwantes D, Jiménez B, et al. 2023. Biocontrol of Botrytis cinerea on grape berries in Chile: Use of registered biofungicides and a new chitosan-based fungicide. Horticulturae. 9(7): 746.Article
- Hirose D, Hobara S, Matsuoka S, Kato K, Tanabe Y, et al. 2016. Diversity and community assembly of moss-associated fungi in ice-free coastal outcrops of continental Antarctica. Fungal Ecol. 24: 94–101. Article
- Hirose D, Hobara S, Tanabe Y, Uchida M, Kudoh S, et al. 2017. Abundance, richness, and succession of microfungi in relation to chemical changes in Antarctic moss profiles. Polar Biol. 40(12): 2457–2468. ArticlePDF
- Ismail MA, Amin MA, Eid AM, Hassan SED, Mahgoub HAM, et al. 2021. Comparative study between exogenously applied plant growth hormones versus metabolites of microbial endophytes as plant growth-promoting for Phaseolus vulgaris L. Cells. 10(5): 1059.ArticlePubMedPMC
- Johnston PR, Quijada L, Smith CA, Baral HO, Hosoya T, et al. 2019. A multigene phylogeny toward a new phylogenetic classification of Leotiomycetes. IMA Fungus. 10(1): 1.ArticlePubMedPMCPDF
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, et al. 2012. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12): 1647–1649. ArticlePubMedPMCPDF
- Kendrick B. 2011. Fungi: Ecological importance and impact on humans. In Encyclopedia of Life Sciences. Wiley.
- Khokhar I, Mukhtar I, Mushtaq S. 2011. Antifungal effect of Penicillium metabolites against some fungi. Arch Phytopathol Plant Prot. 44(14): 1347–1351. Article
- Kim BS, Hwang BK. 2007. Microbial fungicides in the control of plant diseases. J Phytopathol. 155(11-12): 641–653. Article
- Kiss L. 2012. Limits of nuclear ribosomal DNA internal transcribed spacer (ITS) sequences as species barcodes for fungi. Proc Natl Acad Sci USA. 109(27): E1811.ArticlePubMedPMC
- Kochkina GA, Ivanushkina NE, Lupachev AV, Starodumova IP, Vasilenko OV, et al. 2019. Diversity of mycelial fungi in natural and human-affected Antarctic soils. Polar Biol. 42(1): 47–64. ArticlePDF
- Kochkina GA, Ozerskaya SM, Ivanushkina NE, Chigineva NI, Vasilenko OV, et al. 2014. Fungal diversity in the Antarctic active layer. Microbiology. 83(1–2): 94–101. ArticlePDF
- Latorre BA, Elfar K, Ferrada EE. 2015. Gray mold caused by Botrytis cinerea limits grape production in Chile. Cienc Inv Agrar. 42(3): 305–330. Article
- Marx JC, Collins T, D’Amico S, Feller G, Gerday C. 2007. Cold-adapted enzymes from marine Antarctic microorganisms. Mar Biotechnol. 9: 293–304. ArticlePDF
- Mishra PK, Fox RTV, Culham A. 2000. Application of nr‐DNA ITS sequence for identification of Fusarium culmorum isolates. EPPO Bull. 30(3–4): 493–498. Article
- Naranjo‐Ortiz MA, Gabaldón T. 2019. Fungal evolution: Major ecological adaptations and evolutionary transitions. Biol Rev. 94(4): 1443–1476. ArticlePubMedPMCPDF
- Núñez-Montero K, Barrientos L. 2018. Advances in Antarctic research for antimicrobial discovery: A comprehensive narrative review of bacteria from Antarctic environments as potential sources of novel antibiotic compounds against human pathogens and microorganisms of industrial importance. Antibiotics. 7(4): 90.ArticlePubMedPMC
- O’Donnell K, Nirenberg HI, Aoki T, Cigelnik E. 2000. A multigene phylogeny of the Gibberella fujikuroi species complex: Detection of additional phylogenetically distinct species. Mycoscience. 41(1): 61–78. Article
- O’Donnell K, Sutton DA, Fothergill A, McCarthy D, Rinaldi MG, et al. 2008. Molecular phylogenetic diversity, multilocus haplotype nomenclature, and in vitro antifungal resistance within the Fusarium solani species complex. J Clin Microbiol. 46(8): 2477–2490. ArticlePubMedPMCPDF
- Ordóñez-Enireb E, Cucalón RV, Cárdenas D, Ordóñez N, Coello S, et al. 2022. Antarctic fungi with antibiotic potential isolated from Fort William Point, Antarctica. Sci Rep. 12(1): 21477.PubMedPMC
- Park CH, Kim KM, Elvebakk A, Kim O, Jeong G, et al. 2015. Algal and fungal diversity in Antarctic lichens. J Eukaryot Microbiol. 62(2): 196–205. ArticlePubMedPDF
- Price MN, Dehal PS, Arkin AP. 2010. FastTree 2-Approximately maximum-likelihood trees for large alignments. PLoS One. 5(3): e9490. ArticlePubMedPMC
- Quandt CA, Bushley KE, Spatafora JW. 2015. The genome of the truffle-parasite Tolypocladium ophioglossoides and the evolution of antifungal peptaibiotics. BMC Genomics. 16(1): 553.ArticlePubMedPMCPDF
- Quandt CA, Haelewaters D. 2021. Phylogenetic advances in Leotiomycetes, an understudied clade of taxonomically and ecologically diverse fungi. In Encyclopedia of Mycology, pp. 284–294, Elsevier.
- Rämä T, Hassett BT, Bubnova E. 2017. Arctic marine fungi: From filaments and flagella to operational taxonomic units and beyond. Bot Mar. 60(4): 433–452.
- Ramasamy KP, Mahawar L, Rajasabapathy R, Rajeshwari K, Miceli C, et al. 2023. Comprehensive insights on environmental adaptation strategies in Antarctic bacteria and biotechnological applications of cold-adapted molecules. Front Microbiol. 14: 1197797.ArticlePubMedPMC
- Rastegari AA, Yadav AN, Yadav N. 2020. New and Future Developments in Microbial Biotechnology and Bioengineering: Trends of Microbial Biotechnology for Sustainable Agriculture and Biomedicine Systems: Diversity and Functional Perspectives, Elsevier.
- Roca-Couso R, Flores-Félix JD, Rivas R. 2021. Mechanisms of action of microbial biocontrol agents against Botrytis cinerea. J Fungi. 7(12): 1045.ArticlePubMedPMC
- Rosa LH, Da Costa Coelho L, Pinto OHB, Carvalho-Silva M, Convey P, et al. 2021. Ecological succession of fungal and bacterial communities in Antarctic mosses affected by a fairy ring disease. Extremophiles. 25(5–6): 471–481. ArticlePubMedPDF
- Rosa LH, De Sousa JRP, De Menezes GCA, Da Costa Coelho L, Carvalho-Silva M, et al. 2020. Opportunistic fungi found in fairy rings are present on different moss species in the Antarctic Peninsula. Polar Biol. 43(5): 587–596. ArticlePDF
- Rosa LH, Zani CL, Cantrell CL, Duke SO, Van Dijck P, et al. 2019. Fungi in Antarctica: Diversity, ecology, effects of climate change, and bioprospection for bioactive compounds. In Rosa LH. (ed.), Fungi of Antarctica, pp. 1–17, Springer.
- Scherm B, Balmas V, Spanu F, Pani G, Delogu G, et al. 2013. Fusarium culmorum: Causal agent of foot and root rot and head blight on wheat. Mol Plant Pathol. 14(4): 323–341. PubMed
- Schoch CL, Seifert KA, Huhndorf S, Robert V, Spouge JL, et al. 2012. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for fungi. Proc Natl Acad Sci USA. 109(16): 6241–6246. PubMedPMC
- Schueffler A, Anke T. 2014. Fungal natural products in research and development. Nat Prod Rep. 31(10): 1425–1448. ArticlePubMed
- Shi T, Li XQ, Wang ZM, Zheng L, Yu YY, et al. 2022. Bioactivity-guided screening of antimicrobial secondary metabolites from Antarctic cultivable fungus Acrostalagmus luteoalbus CH-6 combined with molecular networking. Mar Drugs. 20(5): 334.ArticlePubMedPMC
- Shi T, Li XQ, Zheng L, Zhang YH, Dai JJ, et al. 2021. Sesquiterpenoids from the Antarctic fungus Pseudogymnoascus sp. HSX2#-11. Front Microbiol. 12: 688202.ArticlePubMedPMC
- Stamatakis A. 2014. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9): 1312–1313. ArticlePubMedPMCPDF
- Suarez MB, Walsh K, Boonham N, O’Neill T, Pearson S, et al. 2005. Development of real-time PCR (TaqMan®) assays for the detection and quantification of Botrytis cinerea in planta. Plant Physiol Biochem. 43(9): 890–899. ArticlePubMed
- Thambugala KM, Daranagama DA, Phillips AJL, Kannangara SD, Promputtha I, et al. 2020. Fungi vs. fungi in biocontrol: An overview of fungal antagonists applied against fungal plant pathogens. Front Cell Infect Microbiol. 10: 604923.ArticlePubMedPMC
- Varrella S, Barone G, Tangherlini M, Rastelli E, Dell’Anno A, et al. 2021. Diversity, ecological role and biotechnological potential of Antarctic marine fungi. J Fungi. 7(5): 391.ArticlePubMedPMC
- Vicente MF, Basilio A, Cabello A, Peláez F. 2003. Microbial natural products as a source of antifungals. Clin Microbiol Infect. 9(1): 15–32. ArticlePubMed
- Vieira G, Purić J, Morão LG, Dos Santos JA, Inforsato FJ, et al. 2018. Terrestrial and marine Antarctic fungi extracts active against Xanthomonas citri subsp. citri. Lett Appl Microbiol. 67(1): 64–71. ArticlePubMedPDF
- Vinale F, Sivasithamparam K, Ghisalberti EL, Woo SL, Nigro M, et al. 2014. Trichoderma secondary metabolites active on plants and fungal pathogens. Open Mycol J. 8(1): 127–139. Article
- Visagie CM, Houbraken J, Frisvad JC, Hong SB, Klaassen CHW, et al. 2014. Identification and nomenclature of the genus Penicillium. Stud Mycol. 78(1): 343–371. ArticlePubMedPMC
- Wang J, Liu M, Mao C, Li S, Zhou J, et al. 2023. Comparative proteomics reveals the mechanism of cyclosporine production and mycelial growth in Tolypocladium inflatum affected by different carbon sources. Front Microbiol. 14: 1259101.ArticlePubMedPMC
- Wang X, Radwan MM, Taráwneh AH, Gao J, Wedge DE, et al. 2013. Antifungal activity against plant pathogens of metabolites from the endophytic fungus Cladosporium cladosporioides. J Agric Food Chem. 61(19): 4551–4555. ArticlePubMedPMC
- White TJ, Bruns T, Lee S, Taylor J. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols, pp. 315–322, Elsevier.
- Yin Y, Miao J, Shao W, Liu X, Zhao Y, et al. 2023. Fungicide resistance: Progress in understanding mechanism, monitoring, and management. Phytopathology. 113(4): 707–718. ArticlePubMed
- Yu NH, Park SY, Kim JA, Park CH, Jeong MH, et al. 2018. Endophytic and endolichenic fungal diversity in maritime Antarctica based on cultured material and their evolutionary position among Dikarya. Fungal Syst Evol. 2: 263–272. ArticlePubMedPMC
- Zucconi L, Canini F, Temporiti ME, Tosi S. 2020. Extracellular enzymes and bioactive compounds from Antarctic terrestrial fungi for bioprospecting. Int J Environ Res Public Health. 17(18): 6459.ArticlePubMedPMC
Citations
Citations to this article as recorded by

- A Drought-Activated Bacterial Symbiont Enhances Legume Resilience Through Coordinated Amino Acid Metabolism
Susmita Das Nishu, Jee Hyun No, Gui Nam Wee, Tae Kwon Lee
Microorganisms.2026; 14(1): 114. CrossRef - Agaricales from Antarctica: Diversity of basidiomata, research challenges, and future perspectives in polar environments
Fernando Augusto Bertazzo-Silva, Jair Putzke
Fungal Biology Reviews.2025; 54: 100458. CrossRef - Diversity, geographical distribution and environmental adaptations of snow molds
Tamotsu Hoshino
Mycoscience.2025; 66(6): 334. CrossRef






 MSK
MSK

 ePub Link
ePub Link Cite this Article
Cite this Article





