Articles
- Page Path
- HOME > J. Microbiol > Volume 63(5); 2025 > Article
-
Full article
Microbiome therapeutic PMC72 through reverse translational research in gout - Mohammed Solayman Hossain1,2,†, Hoonhee Seo2,†, Kyung-Ann Lee3,†, Asad ul-Haq3, Sukyung Kim2, Sujin Jo1,2, Md Abdur Rahim2, Hanieh Tajdozian1,2, Fatemeh Ghorbanian1,2, Youjin Yoon1,2, Indrajeet Barman1,2, Md Sarower Hossen Shuvo1,2, Hyun-Sook Kim2,3,*, Ho-Yeon Song1,2,*
-
Journal of Microbiology 2025;63(5):e2501002.
DOI: https://doi.org/10.71150/jm.2501002
Published online: May 27, 2025
1Department of Microbiology and Immunology, School of Medicine, Soonchunhyang University, Cheonan 31151, Republic of Korea
2Human Microbiome Medical Research Center, School of Medicine, Soonchunhyang University, Asan 31538, Republic of Korea
3Division of Rheumatology, Department of Internal Medicine, Soonchunhyang University Seoul Hospital, Seoul 04401, Republic of Korea
- *Correspondence Ho-Yeon Song songmic@sch.ac.kr Hyun-Sook Kim healthyra@schmc.ac.kr
- †These authors contributed equally to this work.
© The Microbiological Society of Korea
This is an Open Access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/4.0) which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
ABSTRACT
- Gout is an inflammatory arthritis resulting from the deposition of monosodium urate crystals. Urate-lowering therapies for gout have limitations, including side effects and limited efficacy, highlighting the need for novel therapeutic approaches to improve patient outcomes. In this context, our research team conducted a microbiome analysis of fecal samples from healthy individuals and gout patients, identifying Bifidobacterium as a key biomarker. Subsequently, we isolated and identified this strain, B. longum PMC72, and demonstrated its efficacy in a gout mouse model. In potassium oxonate (PO)-induced hyperuricemia mice, PMC72 significantly alleviated nausea, gait disturbances, ankle inflammation, and improved renal health. These effects were associated with marked reductions in oxidative stress markers, including serum uric acid, blood urea nitrogen, hepatic xanthine oxidase, and malondialdehyde (MDA) levels in serum, liver, and joint samples, as well as the downregulation of inflammation and uric acid transport-related gene expression in kidney samples. These benefits were comparable to those treated with Febuxostat, a standard urate-lowering therapy for gout. Furthermore, gut microbiome analysis revealed that PMC72 restored dysbiosis induced by hyperuricemia, contrasting with the reduced microbial diversity observed with febuxostat alone, and showed a complete recovery to eubiosis when combined with Febuxostat. These findings position PMC72 as a promising microbial therapeutic candidate for gout management, demonstrating significant development potential and serving as a benchmark for reverse translational microbiome-based therapeutic research.
Introduction
Materials and Methods
Results
Discussion
Conclusion
Acknowledgments
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korean government (MSIT) (No. RS-2023-00219563). The Basic Science Research Program supported this research through the National Research Foundation of Korea (NRF), funded by the Ministry of Education (2020R1I1A-3072680). The Soonchunhyang University Research Fund also supported this research.
Additionally, we would like to extend our gratitude to Soonchunhyang University Hospital in Seoul and all the researchers at HM∙MRC for their support in this study. Special thanks also go to Dr. Uhjin Song, M.D., for his medical advice and assistance with editing this manuscript in English.
Conflict of Interest
The authors have no conflicts of interest relevant to this study to disclose.
Ethical Statements
This study was conducted following the Declaration of Helsinki. It was approved by the Institutional Review Board (IRB) for Human Research of Soonchunhyang University Seoul Hospital (IRB number: SCH 2019-12-004). Written consent was obtained from all participants. Ethical approval for animal experiments was obtained from Soonchunhyang University (No B230703).
Supplementary Information
Table S1.
Table S2.
Table S3.
Fig. S1.
Fig. S2.
- Acharya C, Sharma A, Kantharia N. 2015. Involvement of oxidative stress in patients of gout and antioxidant effect of allopurinol. Int J Med Sci Public Health. 4: 168.Article
- Ahmed MA, Krishna R, Rayad N, Albusaysi S, Mitra A, et al. 2024. Getting the dose right in drug development for rare diseases: barriers and enablers. Clin Pharmacol Ther. 116: 1412–1432. ArticlePubMed
- Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, et al. 2019. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol. 37: 852–857. ArticlePubMed
- Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, et al. 2016. DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods. 13: 581–583. ArticlePubMedPMCPDF
- Campion EW, Glynn RJ, Delabry LO. 1987. Asymptomatic hyperuricemia. Risks and consequences in the normative aging study. Am J Med. 82: 421–426. ArticlePubMed
- Cao J, Liu Q, Hao H, Bu Y, Tian X, et al. 2022. Lactobacillus paracasei X11 ameliorates hyperuricemia and modulates gut microbiota in mice. Front Immunol. 13: 940228.ArticlePubMed
- Castilho TJC, Almeida GHDR, Mello EVSL, Campos ACL. 2023. Metagenomics of microbiota following probiotic supplementation in rats subjected to intestinal anastomosis. Surg Open Sci. 14: 22–30. ArticlePMC
- Chao A, Shen TJ. 2003. Nonparametric estimation of Shannon’s index of diversity when there are unseen species in sample. Environ Ecol Stat. 10: 429–443.Article
- Cheema AK, Maier I, Dowdy T, Wang Y, Singh R, et al. 2016. Chemopreventive metabolites are correlated with a change in intestinal microbiota measured in A-T mice and decreased carcinogenesis. PLoS One. 11: e0151190. ArticlePMC
- Cuesta-Zuluaga J, Mueller NT, Corrales-Agudelo V, Velásquez-Mejía EP, Carmona JA, et al. 2017. Metformin is associated with higher relative abundance of mucin-degrading Akkermansia muciniphila and several short-chain fatty acid-producing microbiota in the gut. Diabetes Care. 40: 54–62. ArticlePubMedPDF
- Dalbeth N, Choi HK, Joosten LAB, Khanna PP, Matsuo H, et al. 2019. Gout. Nat Rev Dis Primers. 5: 69.ArticlePubMedPDF
- Dalbeth N, Gosling AL, Gaffo A, Abhishek A. 2021. Gout. Lancet. 397: 1843–1855. ArticlePubMed
- El-Dein AN, Ezzat A, Aly HF, Younis EA, Awad GA, et al. 2023. Hypouricemic, anti-inflammatory, and antioxidant activities of Lactobacillus-based functional yogurt in induced-arthritic male Wistar rats: Therapeutic and protective potentials. Biocatal Agric Biotechnol. 47: 102597.Article
- FitzGerald JD, Dalbeth N, Mikuls T, Brignardello‐Petersen R, Guyatt G, et al. 2020. 2020 American College of Rheumatology guideline for the management of gout. Arthritis Care Res. 72: 744–760.Article
- Gao X, Lin H, Revanna K, Dong Q. 2017. A Bayesian taxonomic classification method for 16S rRNA gene sequences with improved species-level accuracy. BMC Bioinform. 18: 247.ArticlePDF
- Ghanem N, Woolf E, Lee S, Price J, Ramer-Tait A, et al. 2023. P22-017-23 study protocol: A reverse translational approach to understanding the role of the gut microbiome in blueberry polyphenol bioavailability. Curr Dev Nutr. 7: 101720.Article
- Gliozzi M, Malara N, Muscoli S, Mollace V. 2016. The treatment of hyperuricemia. Int J Cardiol. 213: 23–27. ArticlePubMed
- Gopalakrishnan V, Weiner B, Ford CB, Sellman BR, Hammond SA, et al. 2020. Intervention strategies for microbial therapeutics in cancer immunotherapy. Immuno Oncol Technol. 6: 9–17. Article
- He Q, Mok TN, Sin TH, Yin J, Li S, et al. 2023. Global, regional, and national prevalence of gout from 1990 to 2019: Age-period-cohort analysis with future burden prediction. JMIR Public Health Surveill. 9: e45943. ArticlePubMedPMC
- Hoque KM, Dixon EE, Lewis RM, Allan J, Gamble GD, et al. 2020. The ABCG2 Q141K hyperuricemia and gout associated variant illuminates the physiology of human urate excretion. Nat Commun. 11: 2767.ArticlePubMedPMCPDF
- Hyatt D, Chen GL, LoCascio PF, Land ML, Larimer FW, et al. 2010. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinform. 11: 119.ArticlePDF
- Joossens M, De Preter V, Ballet V, Verbeke K, Rutgeerts P, et al. 2012. Effect of oligofructose-enriched inulin (OF-IN) on bacterial composition and disease activity of patients with Crohn’s disease: results from a double-blinded randomised controlled trial. Gut. 61: 958.ArticlePubMed
- Kuo CF, Grainge MJ, Zhang W, Doherty M. 2015. Global epidemiology of gout: prevalence, incidence and risk factors. Nat Rev Rheumatol. 11: 649–662. ArticlePubMedPDF
- Kuo YW, Hsieh SH, Chen JF, Liu CR, Chen CW, et al. 2021. Lactobacillus reuteri TSR332 and Lactobacillus fermentum TSF331 stabilize serum uric acid levels and prevent hyperuricemia in rats. PeerJ. 9: e11209. ArticlePubMedPMCPDF
- Lawley B, Munro K, Hughes A, Hodgkinson AJ, Prosser CG, et al. 2017. Differentiation of Bifidobacterium longum subspecies longum and infantis by quantitative PCR using functional gene targets. PeerJ. 5: e3375. ArticlePubMedPMC
- Lee TH, Chen JJ, Wu CY, Yang CW, Yang HY. 2021. Hyperuricemia and progression of chronic kidney disease: A review from physiology and pathogenesis to the role of urate-lowering therapy. Diagn. 11: 1674.Article
- Lee JJ, Lee JS, Chung MK, Ahn JK, Choi HJ, et al. 2023. Korean guidelines for the management of gout. J Rheum Dis. 30: 141–150. ArticlePubMedPMC
- Lee KA, Ul-Haq A, Seo H, Jo S, Kim S, et al. 2025. Characteristics of skin microbiome associated with disease severity in systemic sclerosis. J Microbiol. 63: e2409018. ArticlePDF
- Li J, Lin S, Vanhoutte PM, Woo CW, Xu A. 2016. Akkermansia muciniphila protects against atherosclerosis by preventing metabolic endotoxemia-induced inflammation in Apoe−/− mice. Circulation. 133: 2434–2446. ArticlePubMed
- Li M, Yang D, Mei L, Yuan L, Xie A, et al. 2014. Screening and characterization of purine nucleoside degrading lactic acid bacteria isolated from Chinese sauerkraut and evaluation of the serum uric acid lowering effect in hyperuricemic rats. PLoS ONE. 9: e105577. ArticlePubMedPMC
- Louisiana Office of Public Health. 2015. Clostridium. https://ldh.la.gov/assets/oph/Center-PHCH/Center-CH/infectious-epi/EpiManual/ClostridiumManual.pdfPDF
- Lowe TM, Eddy SR. 1997. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25: 955–964. ArticlePubMedPMC
- Maiuolo J, Oppedisano F, Gratteri S, Muscoli C, Mollace V. 2016. Regulation of uric acid metabolism and excretion. Int J Cardiol. 213: 8–14. ArticlePubMed
- Misra S, Raghuwanshi S. 2022. Safety concerns, regulatory guidelines, current market trends, and future directions toward the use of probiotics in gut-brain-skin axis. In Kaur IP, Beri K, Deol PK, Sandhu SK. (eds.), Probiotic research in therapeutics, vol. 3, pp. 245-268, Springer SingaporeArticle
- Miyata H, Takada T, Toyoda Y, Matsuo H, Ichida K, et al. 2016. Identification of febuxostat as a new strong ABCG2 inhibitor: Potential applications and risks in clinical situations. Front Pharmacol. 7: 518.ArticlePubMedPMC
- Nawrocki EP, Burge SW, Bateman A, Daub J, Eberhardt RY, et al. 2015. Rfam 12.0: updates to the RNA families database. Nucleic Acids Res. 43: D130–D137. ArticlePubMed
- Ohno I. 2011. Relationship between hyperuricemia and chronic kidney disease. Nucleosides Nucleotides Nucleic Acids. 30: 1039–1044. ArticlePubMed
- Oliveira EP, Burini RC. 2012. High plasma uric acid concentration: causes and consequences. Diabetol Metab Syndr. 4: 12.ArticlePubMedPMC
- Palygin O, Klemens CA, Isaeva E, Levchenko V, Spires DR, et al. 2021. Characterization of purinergic receptor 2 signaling in podocytes from diabetic kidneys. iScience. 24: 102528.ArticlePubMedPMC
- Patil T, Soni A, Acharya S. 2021. A brief review on in vivo models for Gouty Arthritis. Metab Open. 11: 100100.Article
- Rahal A, Kumar A, Singh V, Yadav B, Tiwari R, et al. 2014. Oxidative stress, prooxidants, and antioxidants: The interplay. BioMed Res Int. 2014: 761264.ArticlePubMedPMCPDF
- Rahim MA, Seo H, Kim S, Barman I, Ghorbanian F, et al. 2024. Exploring the potential of Lactocaseibacillus rhamnosus PMC203 in inducing autophagy to reduce the burden of Mycobacterium tuberculosis. Med Microbiol Immunol. 213: 14.ArticlePMCPDF
- Rezazadeh L, Alipour B, Jafarabadi MA, Behrooz M, Gargari BP. 2021. Daily consumption effects of probiotic yogurt containing Lactobacillus acidophilus La5 and Bifidobacterium lactis Bb12 on oxidative stress in metabolic syndrome patients. Clin Nutr ESPEN. 41: 136–142. Article
- Rochat T, Bermúdez-Humarán L, Gratadoux JJ, Fourage C, Hoebler C, et al. 2007. Anti-inflammatory effects of Lactobacillus casei BL23 producing or not a manganese-dependant catalase on DSS-induced colitis in mice. Microb Cell Fact. 6: 22.ArticlePMCPDF
- Schmittgen TD, Zakrajsek BA, Mills AG, Gorn V, Singer MJ, et al. 2000. Quantitative reverse transcription-polymerase chain reaction to study mRNA decay: Comparison of endpoint and real-time methods. Anal Biochem. 285: 194–204. ArticlePubMed
- Schroeder BO, Birchenough GMH, Ståhlman M, Arike L, Johansson MEV, et al. 2018. Bifidobacteria or fiber protects against diet-induced microbiota-mediated colonic mucus deterioration. Cell Host Microbe. 23: 27–40. ArticlePubMed
- Seo H, Kim S, Beck S, Song HY. 2024. Perspectives on microbiome therapeutics in infectious diseases: A comprehensive approach beyond immunology and microbiology. Cells. 13: 2003.ArticlePubMedPMC
- Shirvani-Rad S, Khatibzade-Nasari N, Ejtahed HS, Larijani B. 2023. Exploring the role of gut microbiota dysbiosis in gout pathogenesis: a systematic review. Front Med. 10: 1163778.Article
- Sivera F, Andres M, Dalbeth N. 2022. A glance into the future of gout. Ther Adv Musculoskelet Dis. 14: 1759720X221114098.ArticlePubMedPDF
- Tang DH, Ye YS, Wang CY, Li ZL, Zheng H, et al. 2017. Potassium oxonate induces acute hyperuricemia in the tree shrew (tupaia belangeri chinensis). Exp Anim. 66: 209–216. ArticlePubMed
- Thaiss CA, Elinav E. 2017. The remedy within: will the microbiome fulfill its therapeutic promise? J Mol Med. 95: 1021–1027. ArticlePubMedPDF
- Tong S, Zhang P, Cheng Q, Chen M, Chen X, et al. 2022. The role of gut microbiota in gout: Is gut microbiota a potential target for gout treatment. Front Cell Infect Microbiol. 12: 1051682.ArticlePMC
- ul-Haq A, Lee KA, Seo H, Kim S, Jo S, et al. 2022. Characteristic alterations of gut microbiota in uncontrolled gout. J Microbiol. 60: 1178–1190. ArticlePubMedPDF
- Verdura S, Cuyàs E, Martin-Castillo B, Menendez JA. 2019. Metformin as an archetype immuno-metabolic adjuvant for cancer immunotherapy. OncoImmunology. 8: e1633235. ArticlePubMedPMCPDF
- Vieira AT, Galvão I, Amaral FA, Teixeira MM, Nicoli JR, et al. 2015. Oral treatment with Bifidobacterium longum 51A reduced inflammation in a murine experimental model of gout. Benef Microbes. 6: 799–806. ArticlePubMed
- Vorbach C, Harrison R, Capecchi MR. 2003. Xanthine oxidoreductase is central to the evolution and function of the innate immune system. Trends Immunol. 24: 512–517. ArticlePubMed
- Walter J, Ley R. 2011. The human gut microbiome: Ecology and recent evolutionary changes. Annu Rev Microbiol. 65: 411–429. ArticlePubMed
- Wu Y, He F, Li Y, Wang H, Shi L, et al. 2017. Effects of Shizhifang on NLRP3 inflammasome activation and renal tubular injury in hyperuricemic rats. Evid Based Complement Alternat Med. 2017: 7674240.ArticlePubMedPMCPDF
- Wu Y, Ye Z, Feng P, Li R, Chen X, et al. 2021. Limosilactobacillus fermentum JL-3 isolated from “Jiangshui” ameliorates hyperuricemia by degrading uric acid. Gut Microbes. 13: e1897211. Article
- Xi Y, Yan J, Li M, Ying S, Shi Z. 2019. Gut microbiota dysbiosis increases the risk of visceral gout in goslings through translocation of gut-derived lipopolysaccharide. Poultry Sci. 98: 5361–5373. ArticlePDF
- Yadav M, Chauhan NS. 2022. Microbiome therapeutics: exploring the present scenario and challenges. Gastroenterol Rep. 10: goab046.ArticlePDF
- Yan Y, Yu L, Chen B, Cao C, Zhao H, et al. 2023. Mastoparan M suppressed NLRP3 inflammasome activation by inhibiting MAPK/NF-κB and oxidative stress in gouty arthritis. J Inflamm Res. 16: 6179–6193. ArticlePubMedPMCPDF
- Yoon SH, Ha S, Lim J, Kwon S, Chun J. 2017. A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie van Leeuwenhoek. 110: 1281–1286. ArticlePubMedPDF
- Zhang L, Liu J, Jin T, Qin N, Ren X, et al. 2022. Live and pasteurized Akkermansia muciniphila attenuate hyperuricemia in mice through modulating uric acid metabolism, inflammation, and gut microbiota. Food Funct. 13: 12412–12425. ArticlePubMed
- Zhang R, Zhan S, Li S, Zhu Z, He J, et al. 2018. Anti-hyperuricemic and nephroprotective effects of extracts from Chaenomeles sinensis (Thouin) Koehne in hyperuricemic mice. Food Funct. 9: 5778–5790. ArticlePubMed
References
Figure & Data
References
Citations

- Characterization of Gut Microbiota of Honey Bees in Korea
Md Sarower Hossen Shuvo, Sukyung Kim, Sujin Jo, Md Abdur Rahim, Indrajeet Barman, Mohammed Solayman Hossain, Yoonkyoung Jeong, Hwasik Jeong, Sangrim Kim, Hoonhee Seo, Ho-Yeon Song
Polish Journal of Microbiology.2025; 74(4): 428. CrossRef - Quantitative assessment of microbial dynamics in livestock manure and municipal wastewater treatment plants
Geon Choi, Hokyung Song, Tatsuya Unno
Applied Biological Chemistry.2025;[Epub] CrossRef - Flavonifractor plautii as a Next-Generation Probiotic Enhancing the NGP F/P Index in a Simulated Human Gut Microbiome Ecosystem
Md Sarower Hossen Shuvo, Sukyung Kim, Sujin Jo, Md Abdur Rahim, Indrajeet Barman, Mohammed Solayman Hossain, Youjin Yoon, Hanieh Tajdozian, Izaz Ahmed, Ali Atashi, GangWon Jeong, Ho-Seong Suh, JiMin You, Chaemin Sung, Mijung Kim, Hoonhee Seo, Ho-Yeon Song
Pharmaceutics.2025; 17(12): 1603. CrossRef
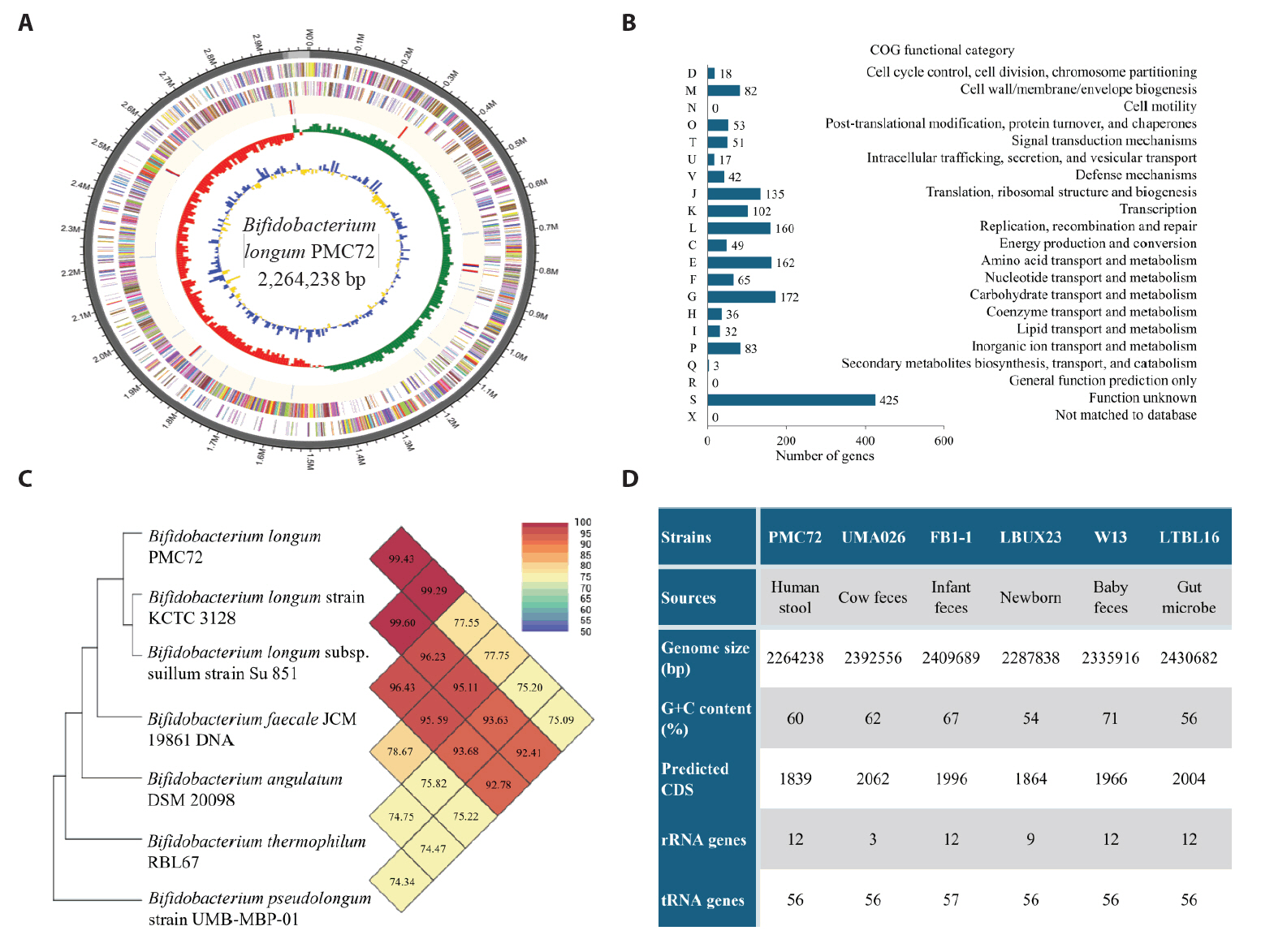
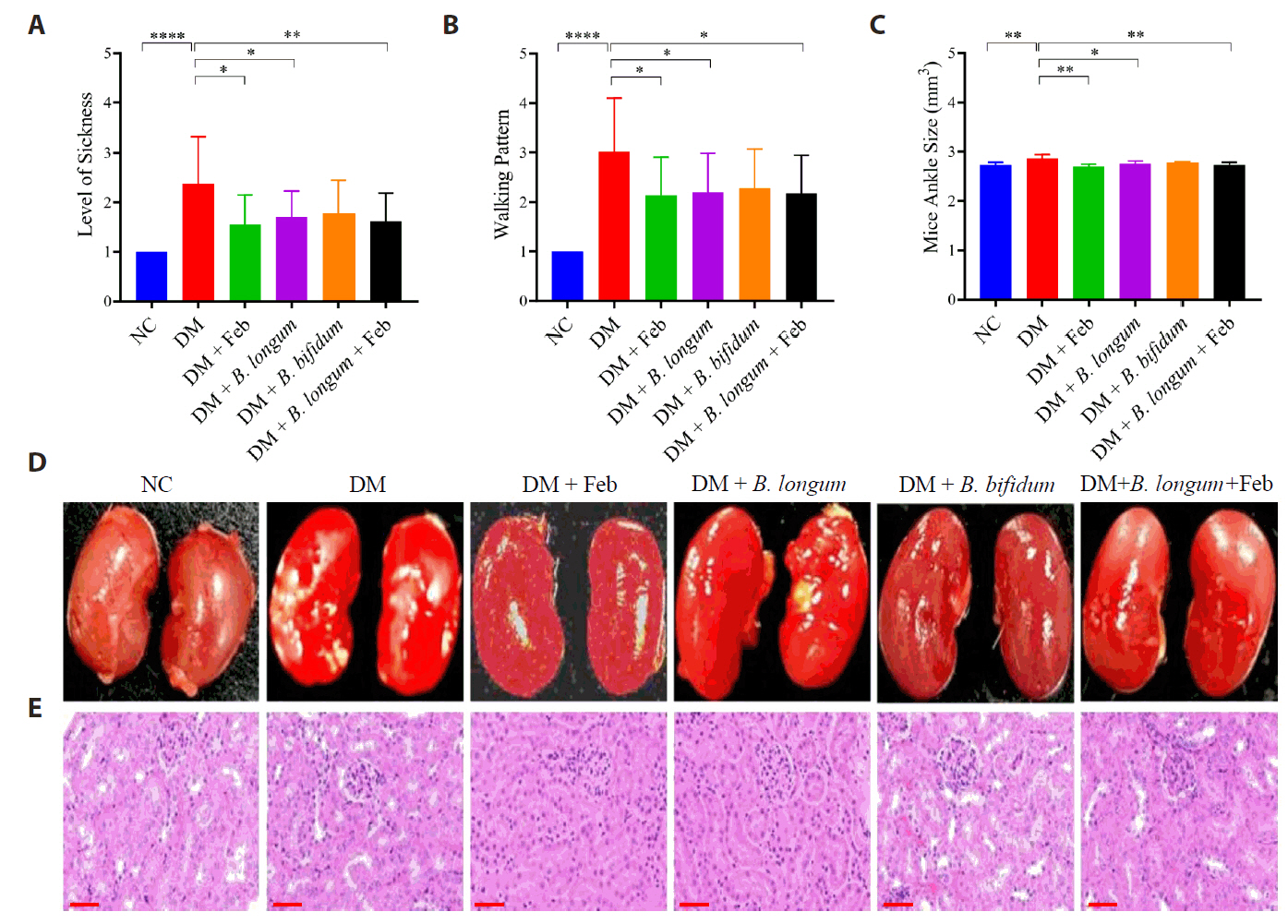
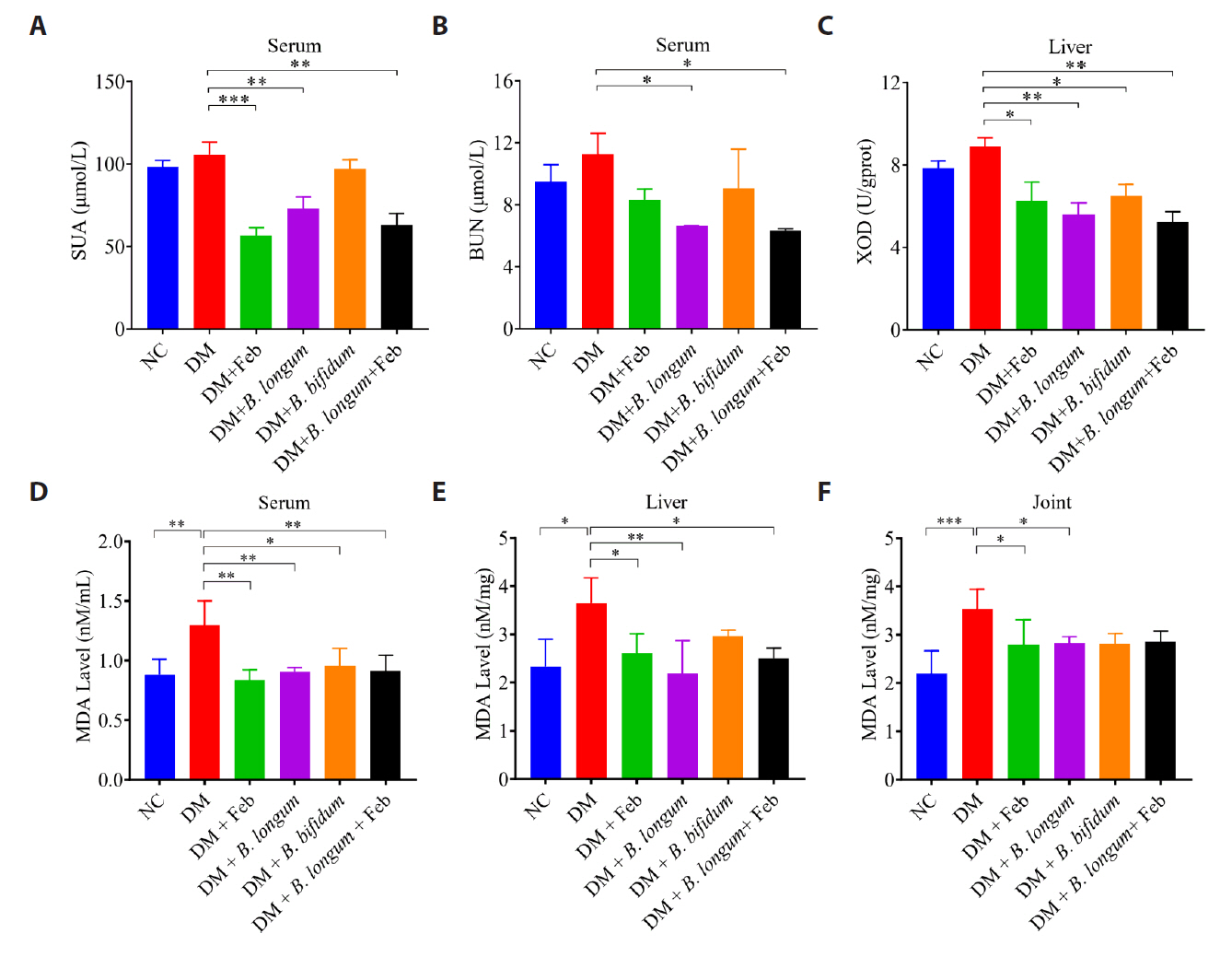
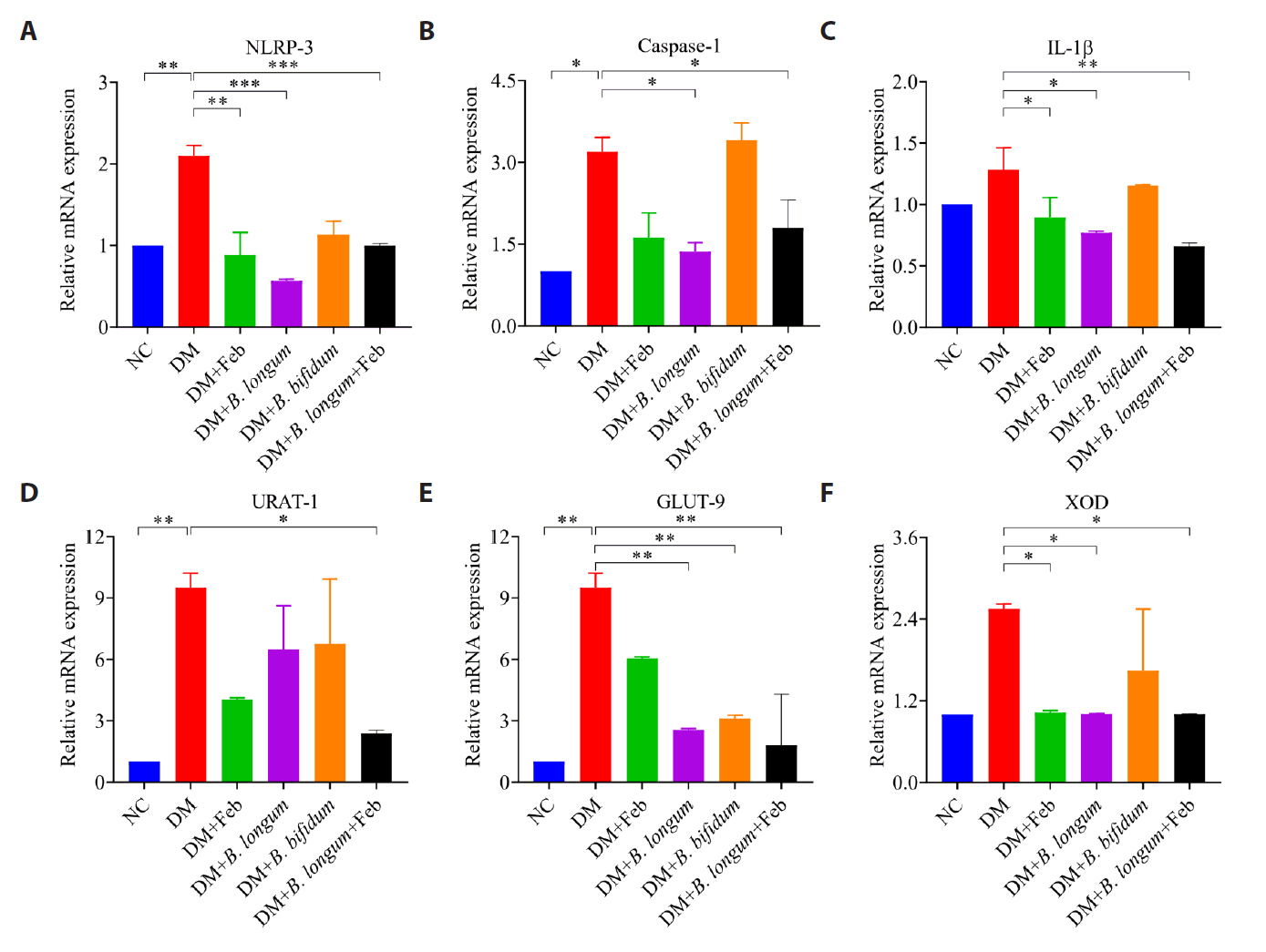
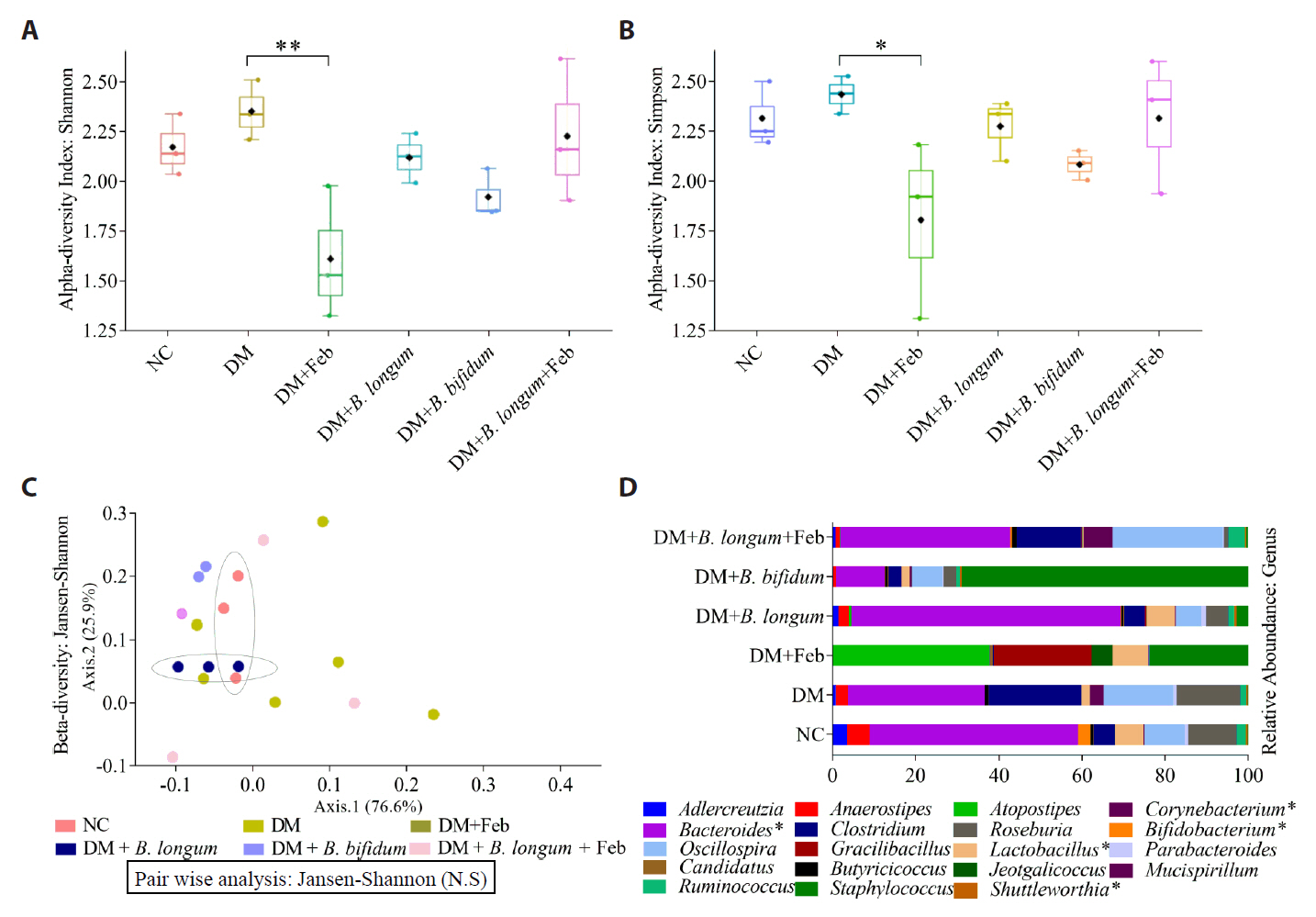
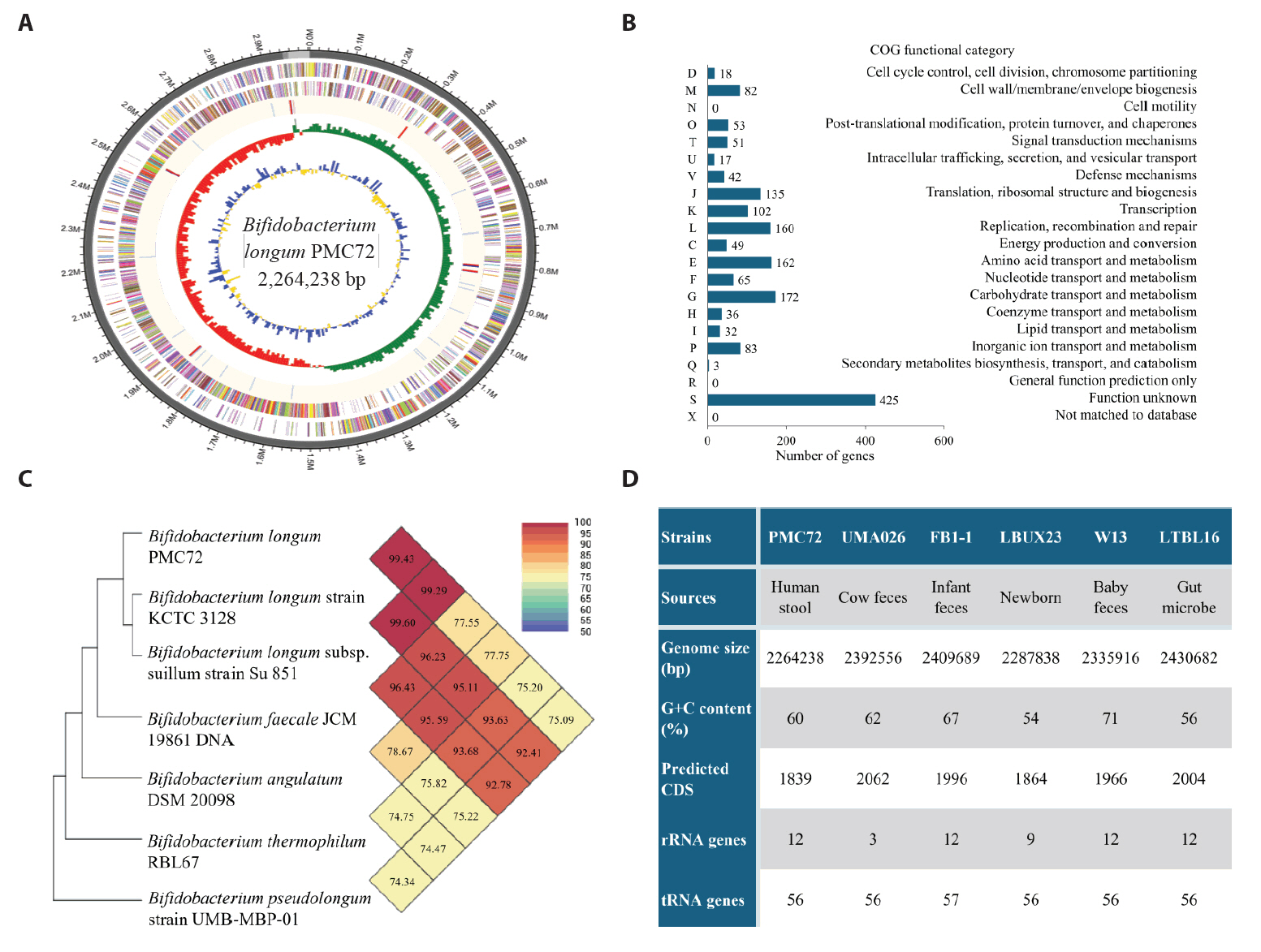
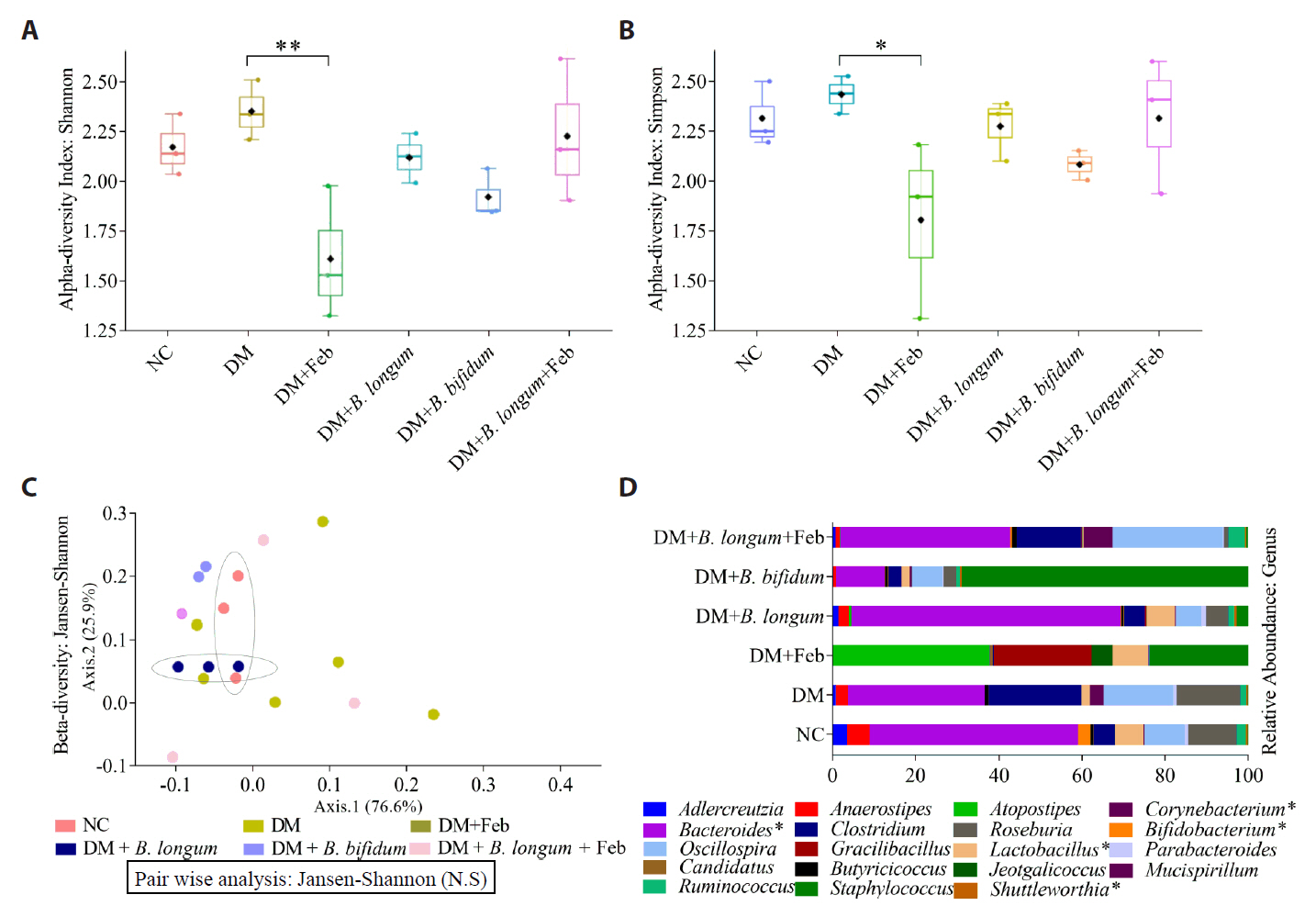
Fig. 1.
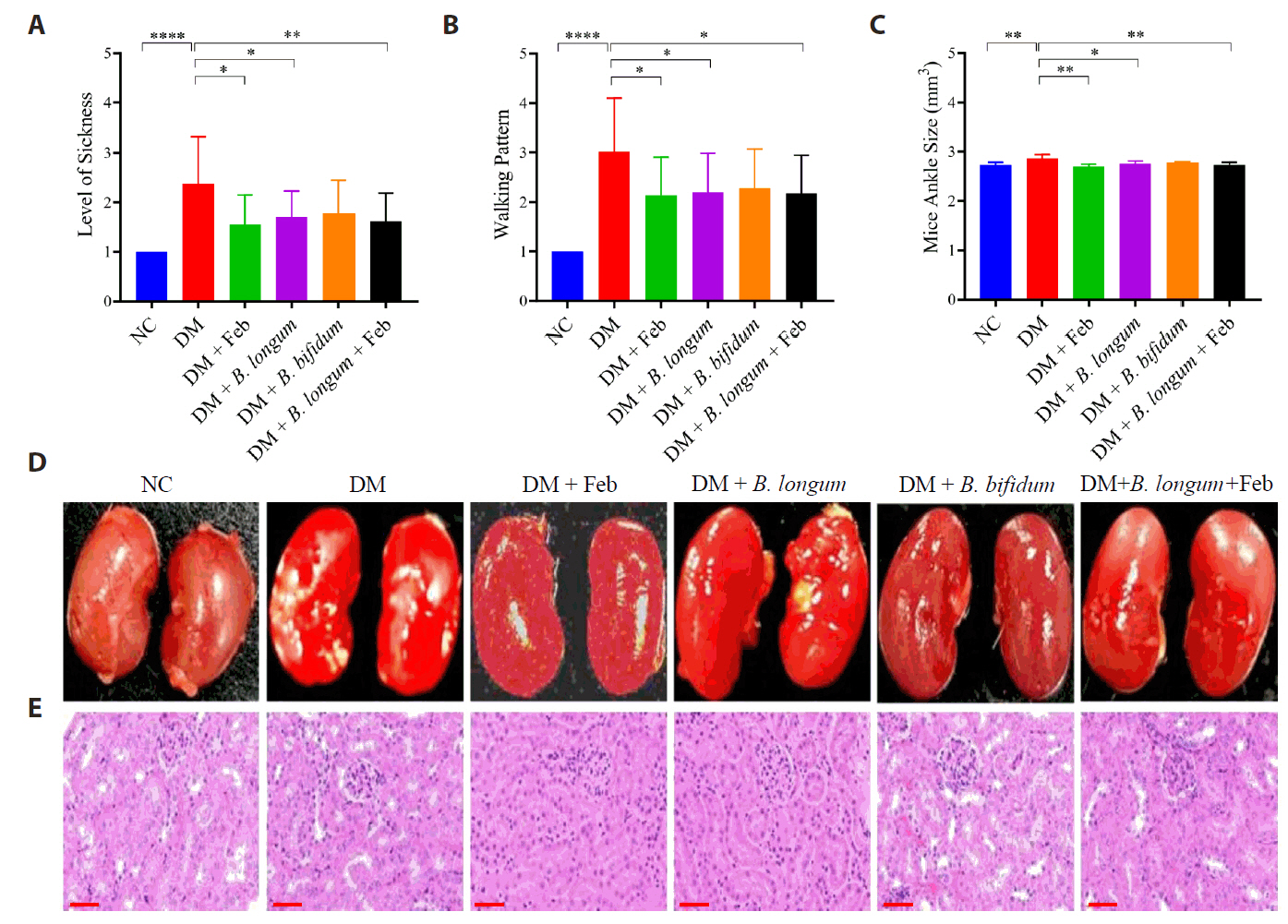
Fig. 2.
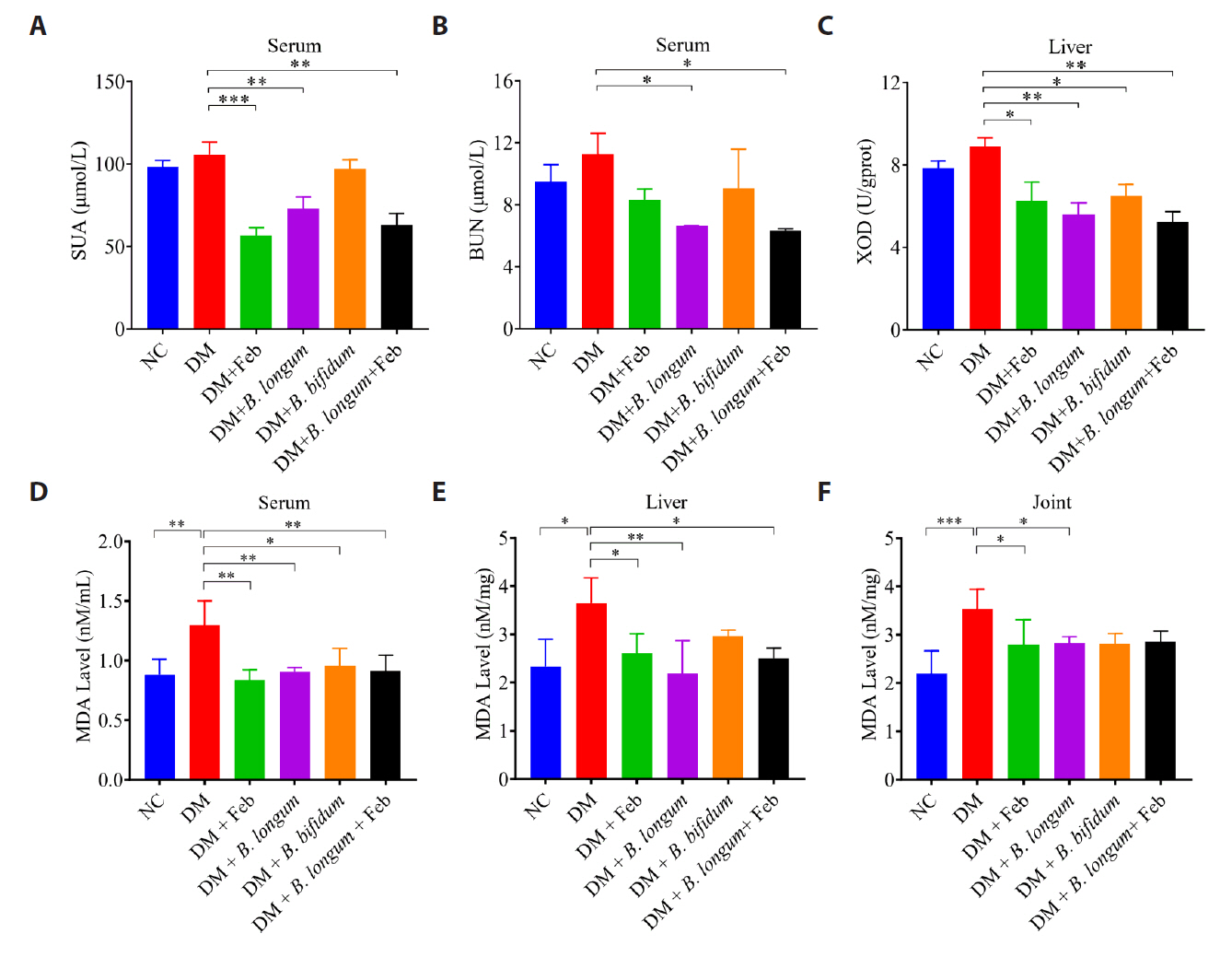
Fig. 3.
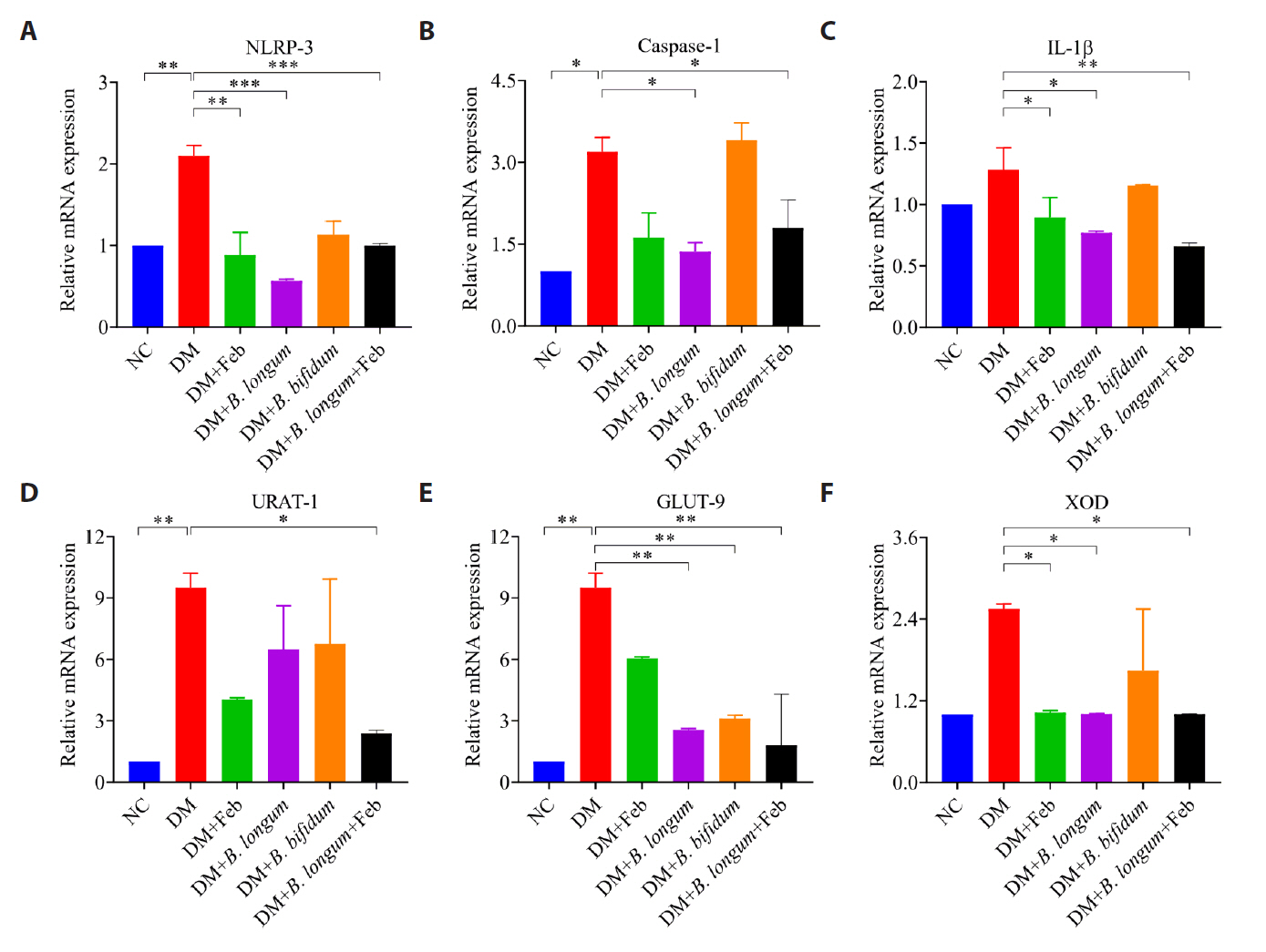
Fig. 4.
Fig. 5.
| NCBI Reference | Organism | Length | Score | Identities |
|---|---|---|---|---|
| NR_145535.1 | Bifidobacterium longum subsp. suillum strain Su 851 | 1428 | 2597 bits (1406) | 1421/1428 (99%) |
| NR_117506.1 | Bifidobacterium longum strain KCTC 3128 | 1488 | 2669 bits (1445) | 1454/1458 (99%) |
| NR_043437.1 | Bifidobacterium longum subsp. infantis strain ATCC 15697 | 1453 | 2662 bits (1441) | 1450/1454 (99%) |
| NR_044691.2 | Bifidobacterium longum subsp. suis strain ATCC 27533 | 1344 | 2460 bits (1332) | 1341/1345 (99%) |
| NR_044693.2 | Bifidobacterium breve DSM 20213 | 1513 | 2361 bits (1278) | 1408/1470 (96%) |
| NR_040783.1 | Bifidobacterium cebidarum strain CCUG 73785 | 1520 | 2361 bits (1278) | 1409/1472 (96%) |
| NR_029137.1 | Bifidobacterium saeculare strain RA161 | 1514 | 2361 bits (1278) | 1397/1455 (96%) |
| NR_036856.1 | Bifidobacterium pullorum strain B 145 | 1517 | 2357 bits (1276) | 1397/1457 (96%) |
| NR_036855.1 | Bifidobacterium gallinarum strain Ch206-5 | 1509 | 2340 bits (1267) | 1395/1459 (96%) |
| NR_037117.1 | Bifidobacterium pseudocatenulatum strain B1279 | 1513 | 2331 bits (1262) | 1395/1459 (96%) |
| NR_037115.2 | Bifidobacterium dentium strain B764 | 1518 | 2329 bits (1261) | 1398/1463 (96%) |
NCBI, National Center for Biotechnology Information
Table 1.
TOP
 MSK
MSK

 ePub Link
ePub Link Cite this Article
Cite this Article






