Articles
- Page Path
- HOME > J. Microbiol > Volume 63(5); 2025 > Article
-
Full article
Characterization of novel bacteriophages for effective phage therapy against Vibrio infections in aquaculture - Kira Moon†,*, Sangdon Ryu†, Seung Hui Song, Se Won Chun, Nakyeong Lee, Aslan Hwanhwi Lee
-
Journal of Microbiology 2025;63(5):e2502009.
DOI: https://doi.org/10.71150/jm.2502009
Published online: May 27, 2025
Division of Environmental Materials, Honam National Institute of Biological Resources, Mokpo 58762, Republic of Korea
- *Correspondence Kira Moon krmoon@inha.ac.kr
- †These authors contributed equally to this work.
© The Microbiological Society of Korea
This is an Open Access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/4.0) which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
ABSTRACT
-
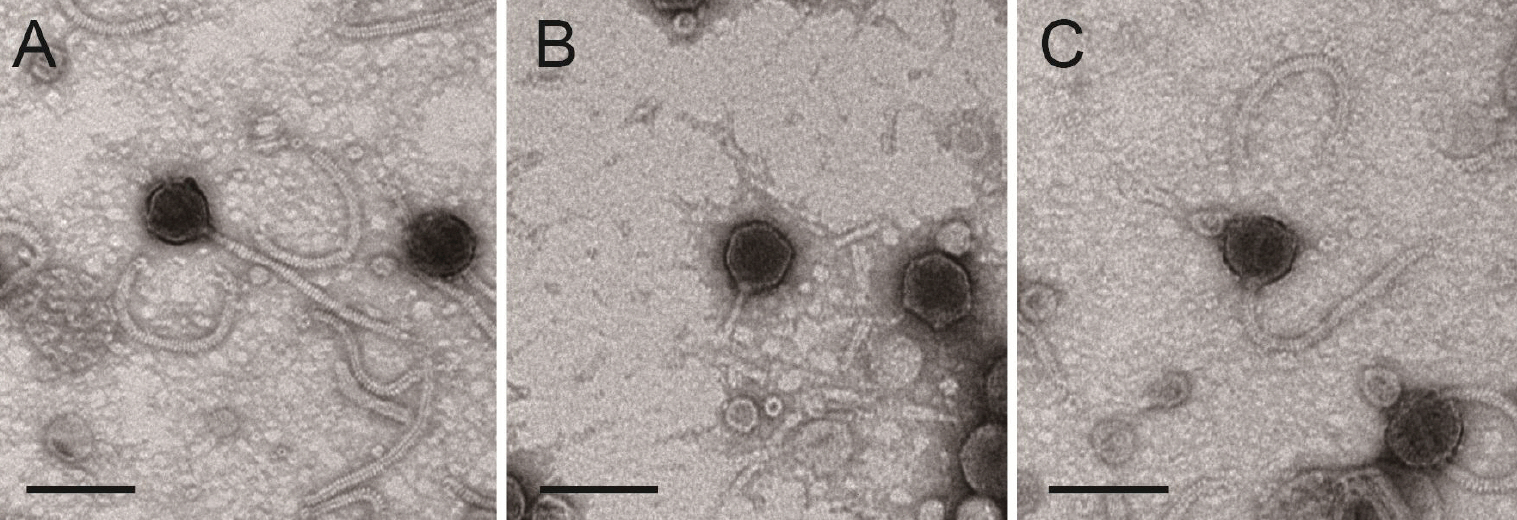
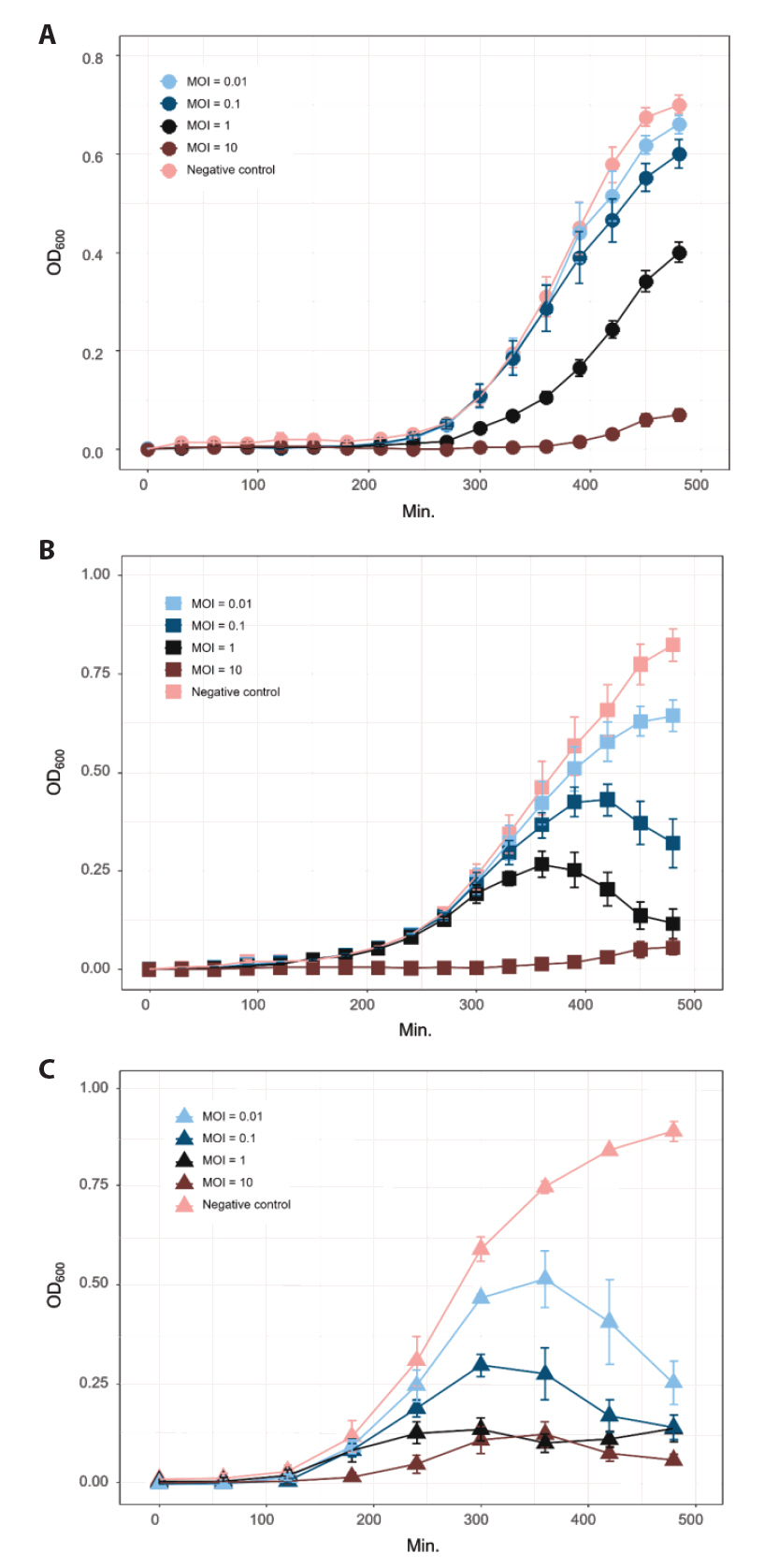
- The widespread use of antibiotics in aquaculture has led to the emergence of multidrug-resistant pathogens and environmental concerns, highlighting the need for sustainable, eco-friendly alternatives. In this study, we isolated and characterized three novel bacteriophages from aquaculture effluents in Korean shrimp farms that target the key Vibrio pathogens, Vibrio harveyi, and Vibrio parahaemolyticus. Bacteriophages were isolated through environmental enrichment and serial purification using double-layer agar assays. Transmission electron microscopy revealed that the phages infecting V. harveyi, designated as vB_VhaS-MS01 and vB_VhaS-MS03, exhibited typical Siphoviridae morphology with long contractile tails and icosahedral heads, whereas the phage isolated from V. parahaemolyticus (vB_VpaP-MS02) displayed Podoviridae characteristics with an icosahedral head and short tail.
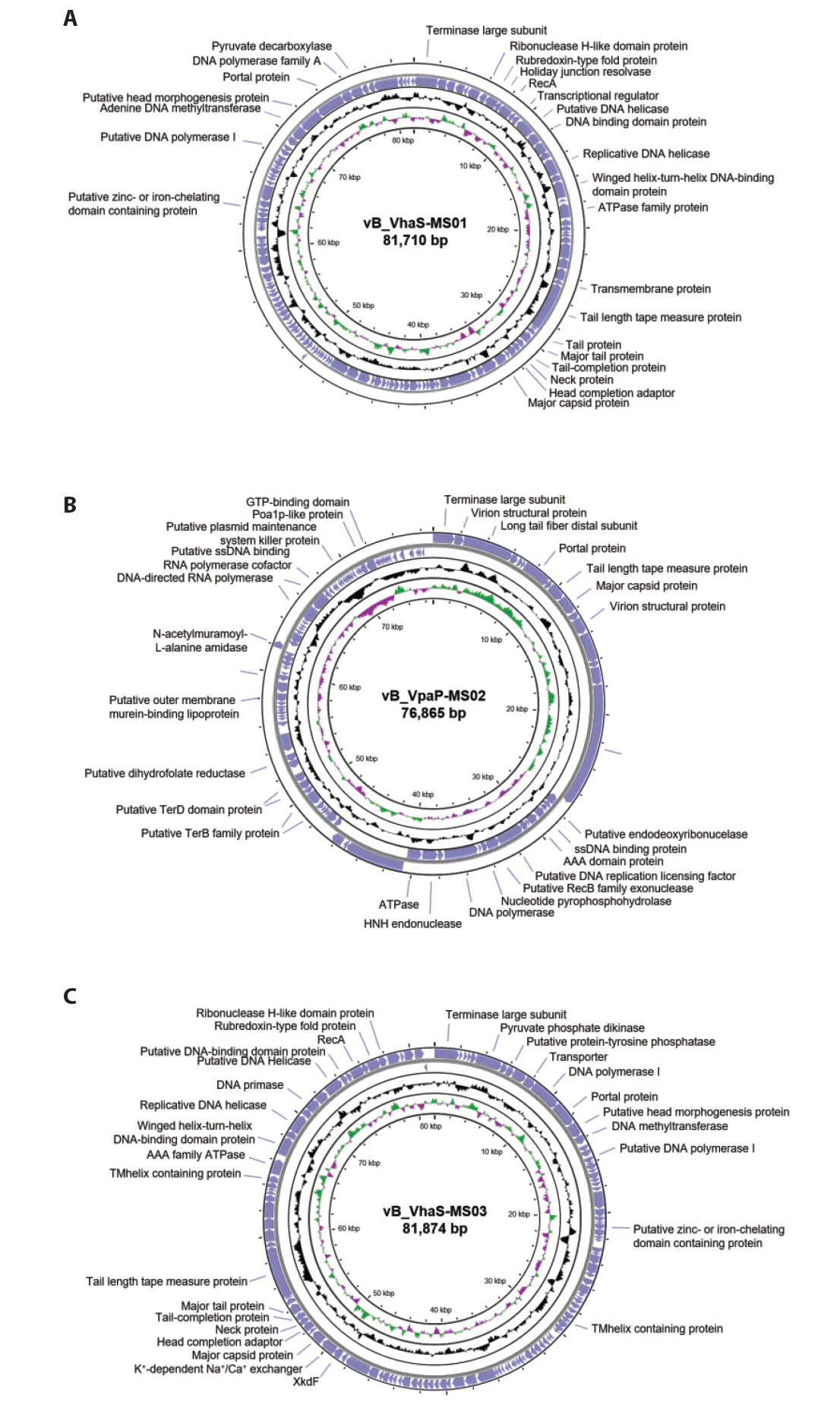
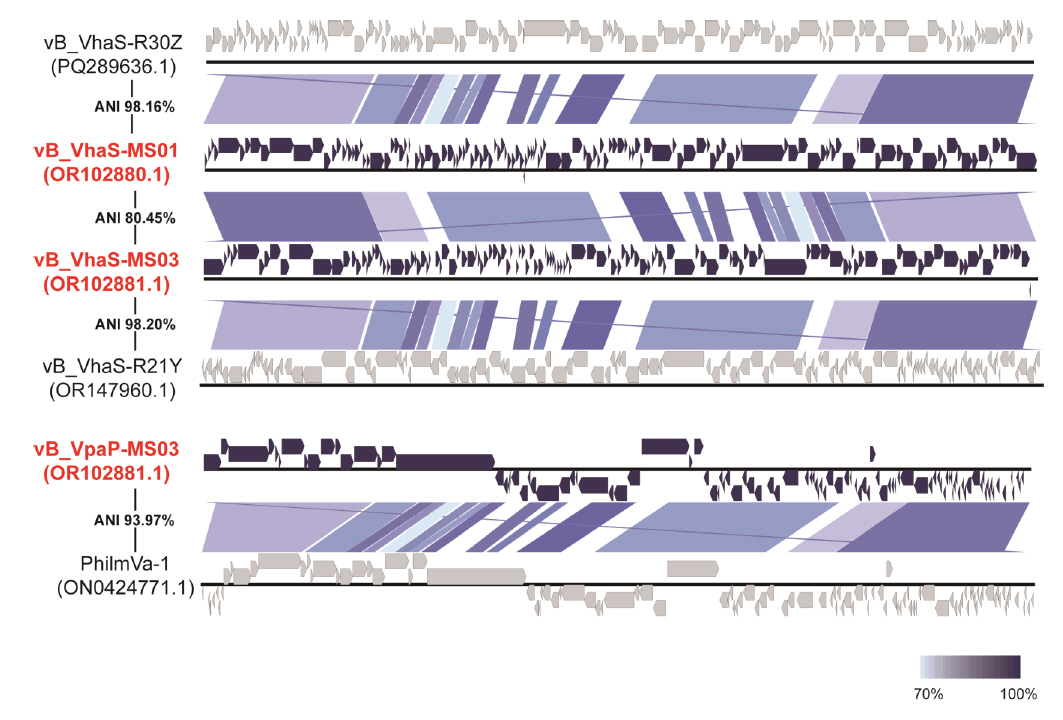
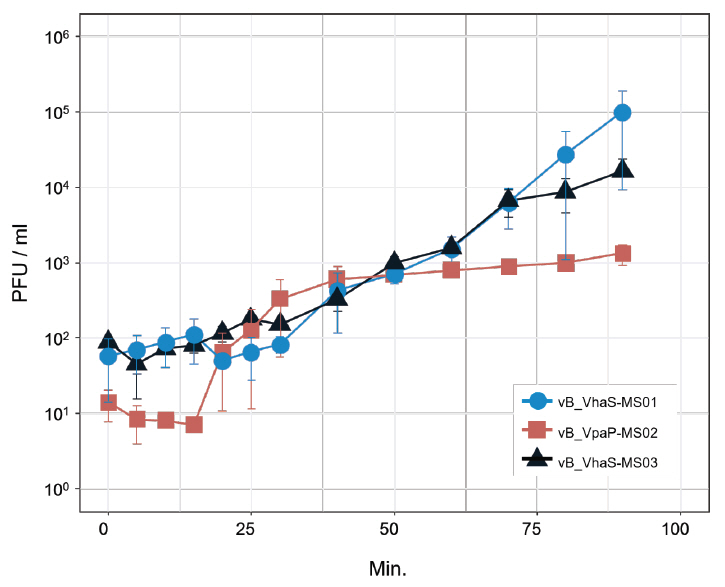
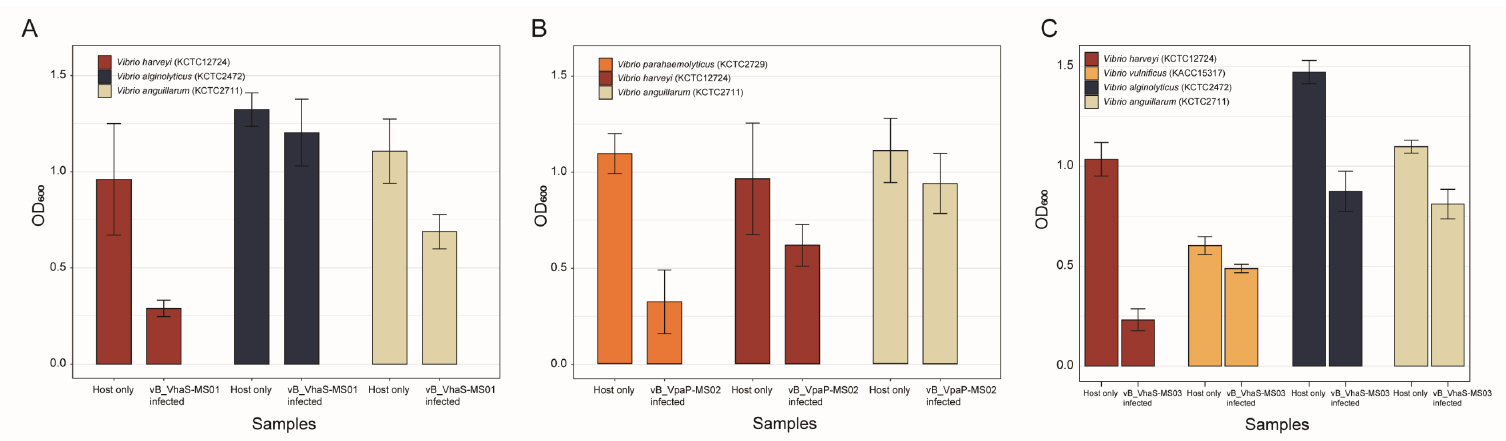
- Whole-genome sequencing produced complete, circularized genomes of 81,710 bp for vB_VhaS-MS01, 81,874 bp for vB_VhaS-MS03, and 76,865 bp for vB_VpaP-MS02, each showing a modular genome organization typical of Caudoviricetes. Genomic and phylogenetic analyses based on the terminase large subunit gene revealed that although vB_VhaS-MS01 and vB_VhaS-MS03 were closely related, vB_VpaP-MS02 exhibited a distinct genomic architecture that reflects its unique morphology and host specificity. Collectively, these comparative analyses demonstrated that all three phages possess genetic sequences markedly different from those of previously reported bacteriophages, thereby establishing their novelty. One-step growth and multiplicity of infection (MOI) experiments demonstrated significant differences in replication kinetics, such as burst size and lytic efficiency, among the phages, with vB_VhaS-MS03 maintaining the most effective bacterial control, even at an MOI of 0.01. Additionally, host range assays showed that vB_VhaS-MS03 possessed a broader spectrum of activity, supporting its potential use as a stand-alone agent or key component of phage cocktails. These findings highlight the potential of region-specific phage therapy as a targeted and sustainable alternative to antibiotics for controlling Vibrio infections in aquaculture.
Introduction
Materials and Methods
Results
Discussion
Accession Numbers
Genome sequences have been submitted to the NCBI and accessible according to following accession numbers: OR102880 (B_VhaS-MS01), OR102882 (vB_VpaP-MS02), and OR102881 (vB_VhaS-MS03).
Acknowledgments
This work was supported by a grant from the Honam National Institute of Biological Resources (HNIBR), funded by the Ministry of Environment (MOE) of the Republic of Korea (HNIBR202302117).
Conflict of Interest
The authors declare that there is no conflict of interest.
Supplementary Information

- Adriaenssens E, Brister JR. 2017. How to name and classify your phage: an informal guide. Viruses. 9: 70.ArticlePubMedPMC
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19: 455–477. ArticlePubMedPMC
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30: 2114–2120. ArticlePubMedPMCPDF
- Cabello FC. 2006. Heavy use of prophylactic antibiotics in aquaculture: a growing problem for human and animal health and for the environment. Environ Microbiol. 8: 1137–1144. ArticlePubMed
- Cai R, Li D, Qin W, Lin W, Pan L, et al. 2024. A novel Vibrio alginolyticus phage and its therapy application in Portunus trituberculatus larvae. Aquaculture. 579: 740165.Article
- Costa P, Pereira C, Romalde JL, Almeida A. 2024. A game of resistance: war between bacteria and phages and how phage cocktails can be the solution. Virology. 110209.ArticlePubMed
- Culot A, Grosset N, Gautier M. 2019. Overcoming the challenges of phage therapy for industrial aquaculture: a review. Aquaculture. 513: 734423.Article
- Director of National Fisheries Quality Management Service. 2024. Acute Hepatopancreatic necrosis Disease Outbreak Alert. National Fisheries Quality Management Service, Ministry of Oceans and Fisheries, Korea. May 27, 2024.
- Doss J, Culbertson K, Hahn D, Camacho J, Barekzi N. 2017. A review of phage therapy against bacterial pathogens of aquatic and terrestrial organisms. Viruses. 9: 50.ArticlePubMedPMC
- Edgar RC. 2022. Muscle5: high-accuracy alignment ensembles enable unbiased assessments of sequence homology and phylogeny. Nat Commun. 13: 6968.ArticlePubMedPMCPDF
- Feng Y, Wei R, Chen Q, Shang T, Zhou N, et al. 2024. Host specificity and cophylogeny in the “animal-gut bacteria-phage” tripartite system. NPJ Biofilms Microbiomes. 10: 72.ArticlePubMedPMCPDF
- Gomes IB, Maillard JY, Simões LC, Simões M. 2020. Emerging contaminants affect the microbiome of water systems—strategies for their mitigation. NPJ Clean Water. 3: 39.ArticlePDF
- Gorbalenya AE, Krupovic M, Mushegian A, Kropinski AM, Siddell SG, et al. 2020. The new scope of virus taxonomy: partitioning the virosphere into 15 hierarchical ranks. Nat Microbiol. 5: 668–674. ArticlePubMedPMCPDF
- Gordillo Altamirano FL, Barr JJ. 2019. Phage therapy in the postantibiotic era. Clin Microbiol Rev. 32: e00066–18. ArticlePubMedPMC
- Grin I, Linke D. 2011. GCView: the genomic context viewer for protein homology searches. Nucleic Acids Res. 39: W353–W356. ArticlePubMedPMC
- Hanson CA, Marston MF, Martiny JBH. 2016. Biogeographic variation in host range phenotypes and taxonomic composition of marine cyanophage isolates. Front Microbiol. 7: 983.ArticlePubMedPMC
- Hill C, Mills S, Ross RP. 2018. Phages & antibiotic resistance: are the most abundant entities on earth ready for a comeback? Future Microbiol. 13: 711–726. ArticlePubMed
- Hossain A, Habibullah-Al-Mamun M, Nagano I, Masunaga S, Kitazawa D, et al. 2022. Antibiotics, antibiotic-resistant bacteria, and resistance genes in aquaculture: risks, current concern, and future thinking. Environ Sci Pollut Res. 29: 11054–11075. ArticlePDF
- Howard-Varona C, Hargreaves KR, Solonenko NE, Markillie LM, White RA 3rd, et al. 2018. Multiple mechanisms drive phage infection efficiency in nearly identical hosts. ISME J. 12: 1605–1618. ArticlePubMedPMCPDF
- Hsu TK, Shih HY, Huang HJ, Hsu JCK, Wang HC, et al. 2024. Isolation and characterization of the novel phage BP14 for lysing Vibrio parahaemolyticus and reducing virulence proteins. Aquaculture. 581: 740484.Article
- Kalatzis PG, Mauritzen JJ, Winther-Have CS, Michniewski S, Millard A, et al. 2023. Staying below the radar: unraveling a new family of ubiquitous “cryptic” non-tailed temperate vibriophages and implications for their bacterial hosts. Int J Mol Sci. 24: 3937.ArticlePubMedPMC
- Kornienko M, Kuptsov N, Gorodnichev R, Bespiatykh D, Guliaev A, et al. 2020. Contribution of Podoviridae and Myoviridae bacteriophages to the effectiveness of anti-staphylococcal therapeutic cocktails. Sci Rep. 10: 18612.ArticlePubMedPMCPDF
- Koskella B, Brockhurst MA. 2014. Bacteria-phage coevolution as a driver of ecological and evolutionary processes in microbial communities. FEMS Microbiol Rev. 38: 916–931. ArticlePubMed
- Koskella B, Meaden S. 2013. Understanding bacteriophage specificity in natural microbial communities. Viruses. 5: 806–823. ArticlePubMedPMC
- Kowalska JD, Kazimierczak J, Sowińska PM, Wójcik EA, Siwicki AK, et al. 2020. Growing trend of fighting infections in aquaculture environment-opportunities and challenges of phage therapy. Antibiotics. 9: 301.ArticlePubMedPMC
- Kropinski AM. 2018. Practical advice on the one-step growth curve. In Clokie MRJ, Kropinski AM, Lavigne R. (eds.), Bacteriophages: Methods and Protocols, Vol. 3, pp. 41–47, Humana Press.Article
- Kumar V, Roy S, Behera BK, Bossier P, Das BK. 2021. Acute hepatopancreatic necrosis disease (AHPND): virulence, pathogenesis and mitigation strategies in shrimp aquaculture. Toxins. 13: 524.ArticlePubMedPMC
- Leptihn S, Loh B. 2022. Complexity, challenges and costs of implementing phage therapy. Future Microbiol. 17: 643–646. ArticlePubMed
- Letchumanan V, Chan KG, Pusparajah P, Saokaew S, Duangjai A, et al. 2016. Insights into bacteriophage application in controlling Vibrio species. Front Microbiol. 7: 1114.ArticlePubMedPMC
- Liu CG, Green SI, Min L, Clark JR, Salazar KC, et al. 2020. Phage-antibiotic synergy is driven by a unique combination of antibacterial mechanism of action and stoichiometry. mBio. 11: 01462–20. Article
- Liu W, Lin YR, Lu MW, Sung PJ, Wang WH, et al. 2014. Genome sequences characterizing five mutations in RNA polymerase and major capsid of phages ϕA318 and ϕAs51 of Vibrio alginolyticus with different burst efficiencies. BMC Genom. 15: 505.ArticlePDF
- Liu J, Wu Q, Xu H, Pan Y, Malakar PK, et al. 2024. Change of antibiotic resistance in Vibrio spp. during shrimp culture in Shanghai. Aquaculture. 580: 740303.Article
- McAllister WT, Raskin CA. 1993. The phage RNA polymerases are related to DNA polymerases and reverse transcriptases. Mol Microbiol. 10: 1–6. ArticlePubMed
- Meyer JR, Dobias DT, Medina SJ, Servilio L, Gupta A, et al. 2016. Ecological speciation of bacteriophage lambda in allopatry and sympatry. Science. 354: 1301–1304. ArticlePubMed
- Moon K, Kang I, Kim S, Kim SJ, Cho JC. 2017. Genome characteristics and environmental distribution of the first phage that infects the LD28 clade, a freshwater methylotrophic bacterial group. Environ Microbiol. 19: 4714–4727. ArticlePubMedPDF
- Nair A, Ghugare GS, Khairnar K. 2022. An appraisal of bacteriophage isolation techniques from environment. Microb Ecol. 83: 519–535. ArticlePubMedPDF
- Nakai T, Park SC. 2002. Bacteriophage therapy of infectious diseases in aquaculture. Res Microbiol. 153: 13–18. ArticlePubMed
- Necel A, Bloch S, Nejman-Faleńczyk B, Dydecka A, Topka-Bielecka G, et al. 2021. A validation system for selection of bacteriophages against shiga toxin-producing Escherichia coli contamination. Toxins. 13: 644.ArticlePubMedPMC
- Nguyen LT, Schmidt HA, Von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32: 268–274. ArticlePubMed
- Park J, Kim B, Song S, Lee YW, Roh E. 2022. Isolation of nine bacteriophages shown effective against Erwinia amylovora in Korea. Plant Pathol J. 38: 248–253. ArticlePubMedPMCPDF
- Piqué N, Miñana-Galbis D, Merino S, Tomás JM. 2015. Virulence factors of Erwinia amylovora: a review. Int J Mol Sci. 16: 12836–12854. ArticlePubMedPMC
- Sanches-Fernandes GMM, Sá-Correia I, Costa R. 2022. Vibriosis outbreaks in aquaculture: addressing environmental and public health concerns and preventive therapies using gilthead seabream farming as a model system. Front Microbiol. 13: 904815.ArticlePubMedPMC
- Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 30: 2068–2069. ArticlePubMedPDF
- Sheikh HI, Alhamadin NII, Liew HJ, Fadhlina A, Wahid MEA, et al. 2024. Virulence factors of the zoonotic pathogen Vibrio alginolyticus: a review and bibliometric analysis. Appl Biochem Microbiol. 60: 514–531. ArticlePDF
- Sieiro C, Areal-Hermida L, Pichardo-Gallardo A, Almuina-Gonzalez R, de Miguel T, et al. 2020. A hundred years of bacteriophage: Can phages replace antibiotics in agriculture and aquaculture? Antibiotics. 9: 493.ArticlePubMedPMC
- Song M, Wu D, Hu Y, Luo H, Li G. 2021. Characterization of an Enterococcus faecalis bacteriophage vB_EfaM_LG1 and its synergistic effect with antibiotic. Front Cell Infect Microbiol. 11: 698807.ArticlePubMedPMC
- Sousa R, Mukherjee S. 2003. T7 RNA polymerase. Prog Nucleic Acid Res Mol Biol. 73: 1–41. ArticlePubMed
- Stone E, Campbell K, Grant I, McAuliffe O. 2019. Understanding and exploiting phage-host interactions. Viruses. 11: 567.ArticlePubMedPMC
- Su H, Duan S, Hu X, Xu W, Xu Y, et al. 2024. Spatiotemporal dynamics, bioaccumulation, and critical influencing factors of antibiotics in tilapia aquaculture: a study on source identification and environmental fate within typical farming systems. J Hazard Mater. 477: 135328.ArticlePubMed
- Sulakvelidze A, Alavidze Z, Morris JG Jr. 2001. Bacteriophage therapy. Antimicrob Agents Chemother. 45: 649–659. ArticlePubMedPMCPDF
- Thompson FL, Iida T, Swings J. 2004. Biodiversity of vibrios. Microbiol Mol Biol Rev. 68: 403–431. ArticlePubMedPMCPDF
- Wick RR, Schultz MB, Zobel J, Holt KE. 2015. Bandage: interactive visualization of de novo genome assemblies. Bioinformatics. 31: 3350–3352. ArticlePubMedPMCPDF
- Yoon SH, Ha SM, Lim J, Kwon S, Chun J. 2017. A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie van Leeuwenhoek. 110: 1281–1286. ArticlePubMedPDF
- Zhang XH, He X, Austin B. 2020. Vibrio harveyi: a serious pathogen of fish and invertebrates in mariculture. Mar Life Sci Technol. 2: 231–245. ArticlePubMedPDF
References
Figure & Data
References
Citations

- Revolutionizing seafood safety with bacteriophages: emerging technologies and applications
Nigar Sultana Meghla, Soo-Jin Jung, Md Furkanur Rahaman Mizan, Syeda Roufun Nesa, IkSoon Kang, Sang-Do Ha
Food Microbiology.2026; 137: 105021. CrossRef - Feed Additives in Aquaculture: Benefits, Risks, and the Need for Robust Regulatory Frameworks
Ekemini Okon, Matthew Iyobhebhe, Paul Olatunji, Mary Adeleke, Nelson Matekwe, Reuben Okocha
Fishes.2025; 10(9): 471. CrossRef
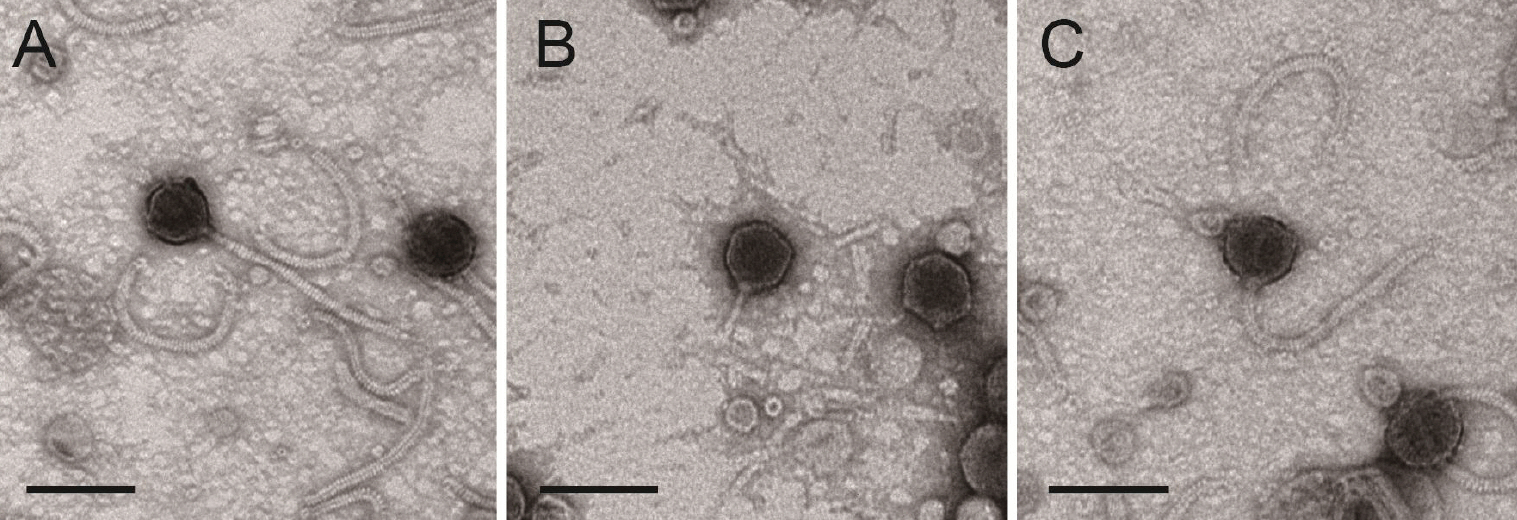
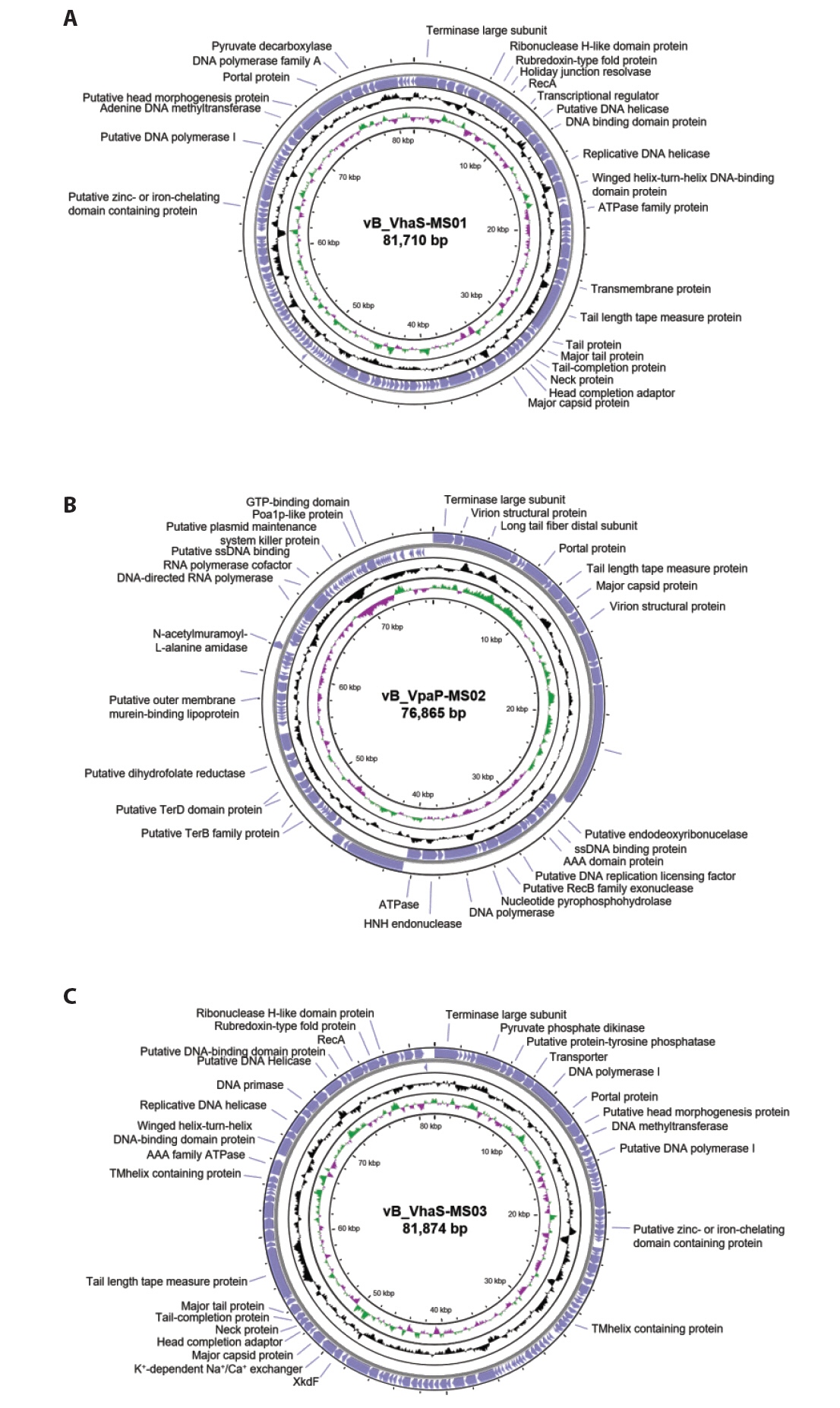

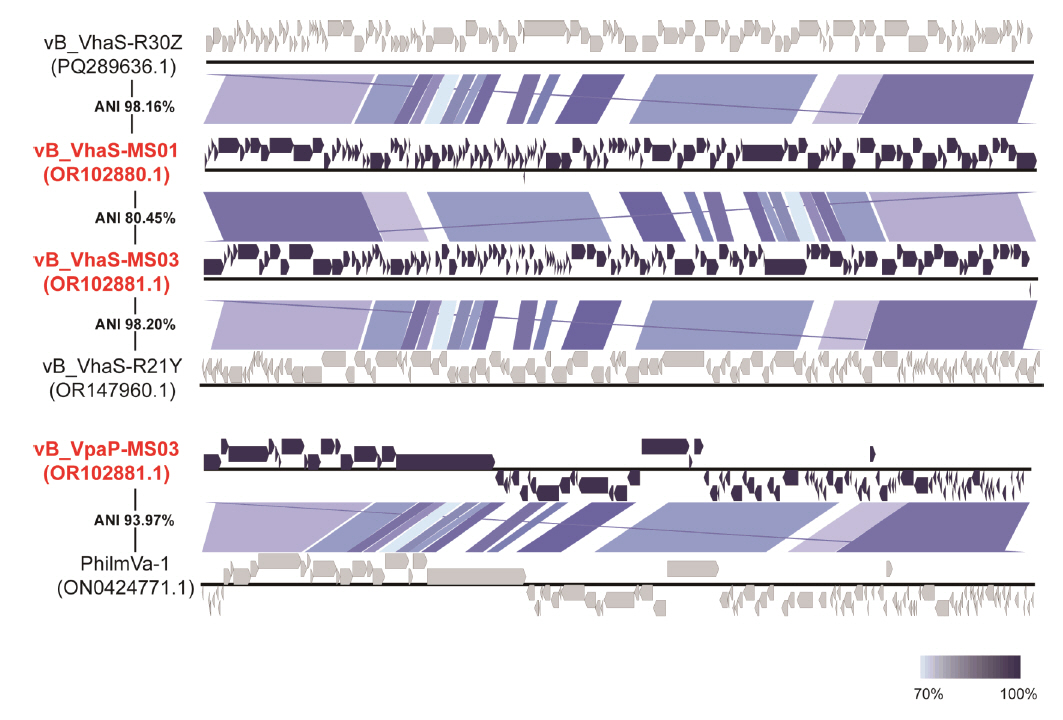
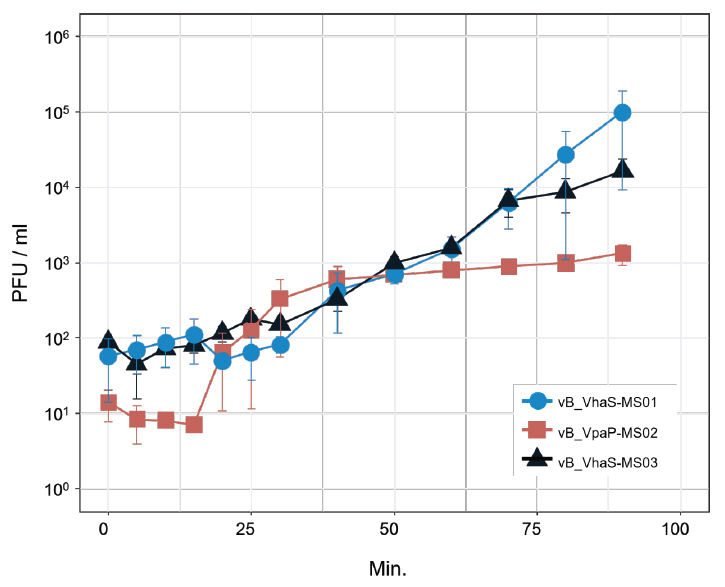
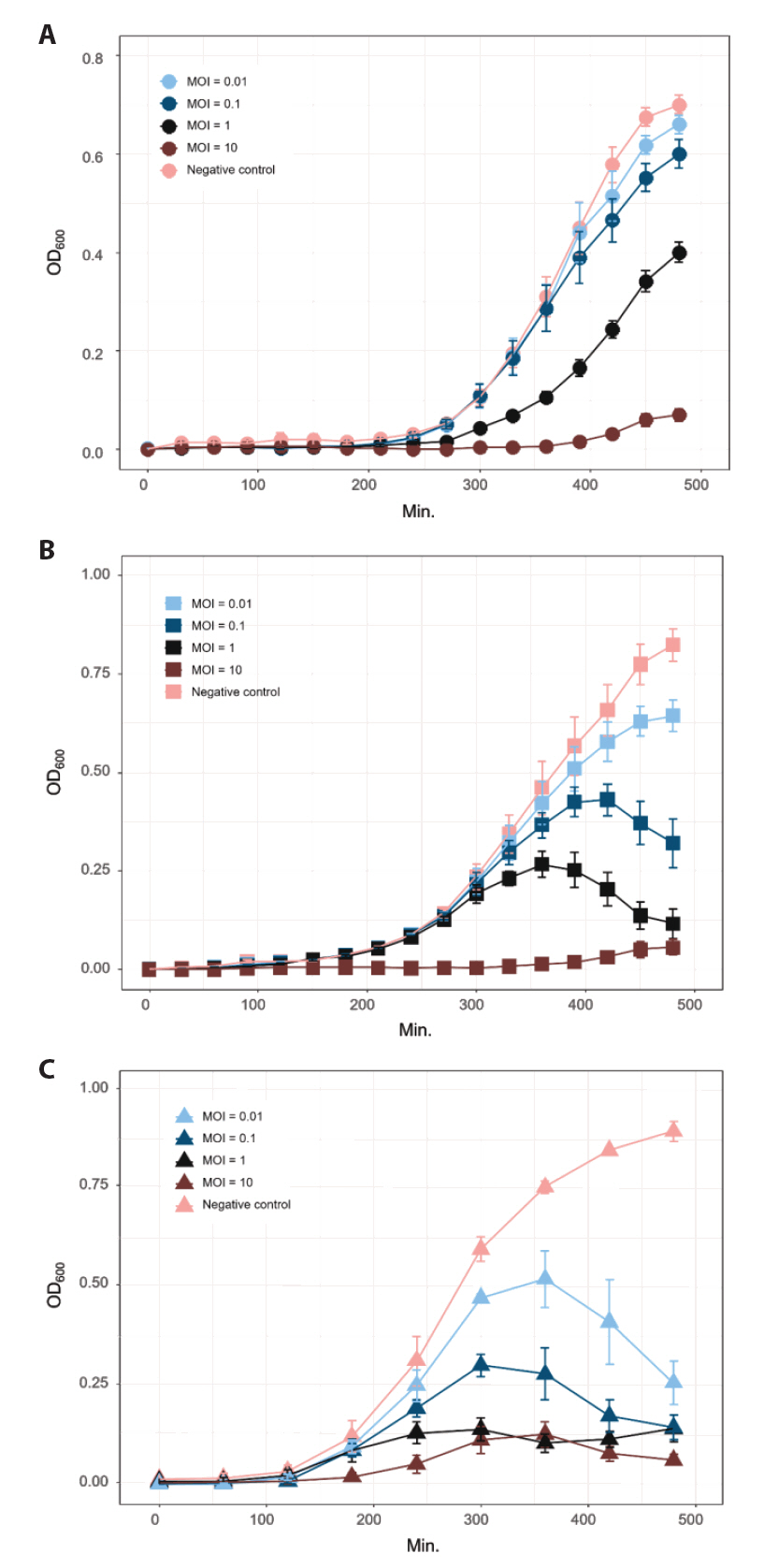
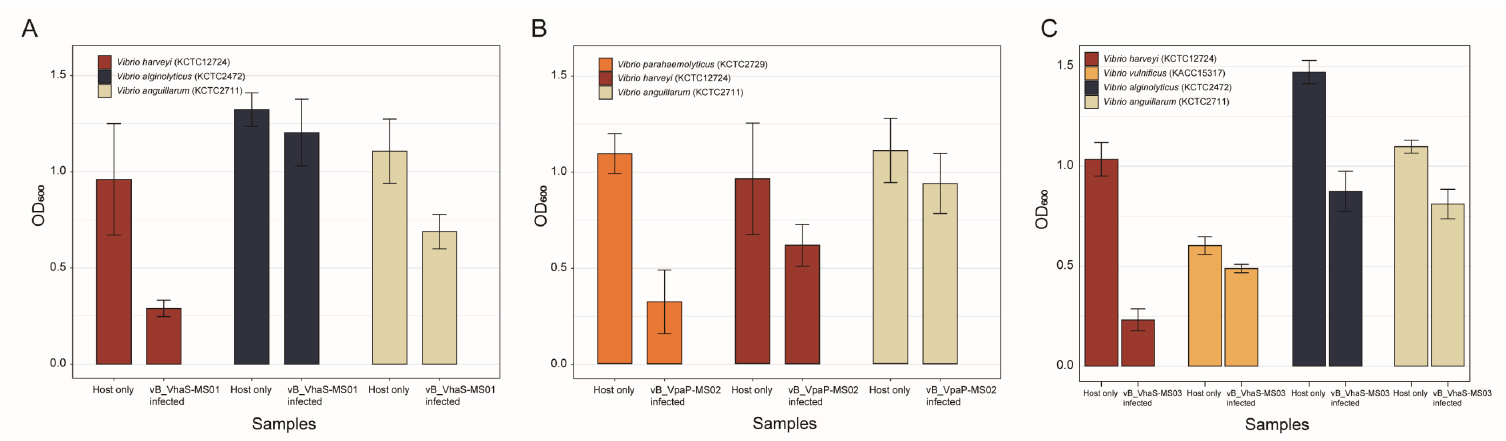
Fig. 1.
Fig. 2.
Fig. 3.
Fig. 4.
Fig. 5.
Fig. 6.
Fig. 7.
| Bacteriophage | vB_VhaS-MS01 | vB_VpaP-MS02 | vB_VhaS-MS03 |
|---|---|---|---|
| Bacterial host | Vibrio harveyi | Vibrio parahaemolyticus | Vibrio harveyi |
| Morphology | Siphoviridae | Podoviridae | Siphoviridae |
| Burst size | 126.38 PFU/ml | 94.76 PFU/ml | 5,525.32 PFU/ml |
| Genome size | 81,710 bp | 76,865 bp | 81,874 bp |
| G+C content | 46.8% | 38.5% | 47.6% |
| No. of CDS | 126 | 102 | 124 |
| Accession no. | OR102880 | OR102882 | OR102881 |
Table 1.
TOP
 MSK
MSK

 ePub Link
ePub Link Cite this Article
Cite this Article








