ABSTRACT
- Two Gram-stain-negative, obligately aerobic, non-motile, short rod-shaped bacteria, designated IMCC43871T and IMCC45206T, were isolated from coastal surface seawater collected from the Yellow Sea and the South Sea of Korea, respectively. The two strains shared 99.2% 16S rRNA gene sequence similarity with each other and exhibited ≤ 98.4% similarity to three described Rubrivirga species. Average nucleotide identity (ANI) and digital DNA–DNA hybridization (dDDH) values between IMCC43871T and IMCC45206T were 88.5% and 36.3%, respectively, confirming that they represent two distinct species. Their ANI (≤ 77.7%) and dDDH (≤ 21.4%) values relative to the type strains of the genus Rubrivirga further supported the recognition of strains IMCC43871T and IMCC45206T as two novel species within the genus. The complete genomes of IMCC43871T (4.17 Mb, 71.8% G + C content) and IMCC45206T (4.17 Mb, 72.8% G + C content) fall within the known genomic range of the genus. Cellular fatty acid, quinone, and polar lipid profiles were consistent with the chemotaxonomic features of the genus Rubrivirga, supporting their affiliation with the genus. Based on phylogenetic, genomic, and phenotypic evidence, strains IMCC43871T and IMCC45206T are proposed as two novel species, Rubrivirga aquatilis sp. nov. and Rubrivirga halophila sp. nov., respectively. The type strains are IMCC43871T (= KCTC 102072T = NBRC 116463T) and IMCC45206T (= KCTC 92925T = NBRC 116172T = CCTCC AB 2023136T).
-
Keywords: Rubrivirga aquatilis, Rubrivirga halophila, polyphasic taxonomy, marine bacteria, novel species, genome
Introduction
The genus Rubrivirga, a member of the family Rubricoccaceae within the class Rhodothermia, currently comprises three validly published species: Rubrivirga marina (Park et al., 2013), the type species; Rubrivirga profundi (Song et al., 2016); and Rubrivirga litoralis (Rey-Velasco et al., 2024). R. marina SAORIC-28T was isolated from deep seawater (3,000 m) in the western Pacific Ocean, followed by R. profundi SAORIC-476T from mesopelagic seawater (500 m) in the same region. The most recently described species, R. litoralis, was obtained from microcosms of coastal surface seawater collected in Blanes Bay, located in the northwestern Mediterranean Sea. Members of the genus Rubrivirga are Gram-stain-negative, non-motile, aerobic or facultatively anaerobic bacteria that exhibit pale red pigmentation and typically grow under mesophilic and slightly halophilic conditions. The genus is characterized by menaquinone-7 (MK-7) as the major respiratory quinone and phosphatidylethanolamine (PE), phosphatidylglycerol (PG), and diphosphatidylglycerol (DPG) as predominant polar lipids. The genome sizes (3.72–4.98 Mb) and DNA G + C contents (71.5–73.7 mol%) of the three described species show a relatively narrow range, indicating genomic coherence within the genus.
Rubrivirga species are known to contribute to marine ecosystems by catabolizing dimethylsulfoniopropionate (DMSP), a key process in sulfur cycling and climate regulation, and by degrading organic matter and detoxifying sulfide in mangrove environments (Rey-Velasco et al., 2024; Wainwright et al., 2023). Notably, R. marina encodes a PL6 family alginate lyase (AlyRm1) with broad substrate specificity and high alkaline tolerance, suggesting ecological adaptation to marine environments and potential for biotechnological applications (Zheng et al., 2023). However, despite the ecological relevance of this genus, only one of the three currently described Rubrivirga species originates from a coastal environment, and their distribution along coastal gradients remains largely unexplored. The Korean coastline, shaped by dynamic estuarine mixing and pronounced seasonal variability, harbors diverse microbial assemblages structured by sharp gradients in salinity, nutrients, and organic matter (Han et al., 2022). In recent years, numerous novel bacterial species have been continuously isolated and described from Korean coastal environments, reflecting the high microbial diversity harbored by these dynamic ecosystems (Lee et al., 2024; Tak et al., 2024; Yang et al., 2023, 2024).
As part of a broader survey of prokaryotic diversity in island-associated marine habitats, two strains, designated IMCC43871T and IMCC45206T, were isolated and characterized from Korean coastal seawater. Comprehensive phenotypic, phylogenetic, chemotaxonomic, and genomic analyses revealed that these strains represent two novel species of the genus Rubrivirga. This finding expands the ecological and taxonomic breadth of the genus Rubrivirga.
Materials and Methods
Isolation and culture condition
Strain IMCC43871T was isolated from coastal surface seawater collected at Daebu Island, Ansan (37°16'58.0"N 126°29'13.0"E) in June 2022, and strain IMCC45206T was obtained from Somaemul Island, Tongyeong (34°37'20.8"N 128°32'55.7"E), Republic of Korea in October 2022. Seawater samples were serially diluted and spread onto marine agar 2216 (MA; Difco). After incubation at 20°C for 5 d, single colonies were streaked and purified through three successive transfers. Following determination of optimal growth temperatures, strains IMCC43871T and IMCC45206T were routinely cultured on MA at 25°C and preserved as 10% (v/v) glycerol suspensions at –80°C. Anaerobic growth was tested using a commercial gas-pack system (AnaeroPack; Mitsubishi Gas Chemical) at 25°C. For comparative phenotypic characterization under identical culture conditions, the closely related type strains R. marina KCTC 23867T and R. profundi KACC 18401T were obtained from the Korean Collection for Type Cultures (KCTC) and the Korean Agricultural Culture Collection (KACC), respectively, and cultivated on MA at 25°C for 7 d.
Phylogenetic analysis based on 16S rRNA gene sequences
Genomic DNA from strains IMCC43871T and IMCC45206T was extracted using the DNeasy PowerSoil Kit (Qiagen) according to the manufacturer's instructions. Amplification of the 16S rRNA genes was performed using the universal bacterial primers 27F and 1492R (Weisburg et al., 1991), and sequencing was conducted by Biofact Co. (Korea) using the Sanger method. The resulting sequences, comprising 1,428 bp for IMCC43871T and 1,455 bp for IMCC45206T, were analyzed using BLASTn against the GenBank database and pairwise sequence comparisons were performed using the EzBioCloud server (Yoon et al., 2017) to determine their phylogenetic affiliations.
For phylogenetic analyses, the 16S rRNA gene sequences of the two strains and their closest relatives were aligned using the SILVA Incremental Aligner and imported into the ARB software package (Ludwig et al., 2004). Aligned sequences were then exported from ARB and used to construct phylogenetic trees with MEGA X (Kumar et al., 2018) to determine the phylogenetic positions of the strains. Three tree-building algorithms were employed: maximum likelihood (ML) based on the Tamura-Nei model (Felsenstein, 1981), neighbor-joining (NJ) using the Jukes-Cantor model (Saitou and Nei, 1987), and minimum-evolution (ME) also using the Jukes-Cantor model (Fitch, 1971). Tree topologies were evaluated by bootstrap analysis with 1,000 replicates (Felsenstein, 1985).
Genome sequencing and analysis
Whole-genome sequencing was performed using the Oxford Nanopore Technologies (ONT) platform with a Flongle flow cell (R9.4.1), following the protocol provided with the Ligation Sequencing Kit (SQK-LSK109). In parallel, short-read sequencing was conducted using a 2 × 150 bp paired-end strategy on the Illumina NovaSeq 6000 platform (DNA Link Corp., Korea). Raw ONT signal data were basecalled using Dorado v7.2.13, and adapter sequences were removed with Porechop v0.2.4. The filtered long reads were assembled using Hybracter with the --medakaModel r941_min_sup_g507 option (Bouras et al., 2024). Genome completeness and contamination were assessed using CheckM v1.1.3 (Parks et al., 2015).
For comparative genomic analyses and assessment of genome relatedness, the genome assemblies of R. marina SAORIC-28T (accession no. MQWD00000000) and R. profundi SAORIC-476T (MVOI00000000) were retrieved from the GenBank database. Average nucleotide identity (ANI) and digital DNA–DNA hybridization (dDDH) values were calculated using JSpeciesWS (Richter et al., 2016) and the Genome-to-Genome Distance Calculator (GGDC 3.0) (Meier-Kolthoff et al., 2013), respectively. To construct a genome-based phylogenetic tree, 81 conserved bacterial core genes were extracted using the UBCG2 pipeline (Kim et al., 2021) and analyzed with RAxML v8.2.12 (Stamatakis, 2014). Functional genome annotation was performed using Prokka (Seemann, 2014), and the predicted protein sequences were assigned to KEGG orthology terms via the BlastKOALA tool (Kanehisa et al., 2016). In addition, the distribution of functional categories based on clusters of orthologous groups (COG) was assessed by querying the protein sequences against the COG database using RPS-BLAST (e-value cutoff: 0.01) (Marchler-Bauer et al., 2013).
Physiological and chemotaxonomic analysis
Cell morphology was observed using transmission electron microscopy (CM200, Philips) with cells negatively stained using 1% (w/v) uranyl acetate on carbon-coated copper grids (Electron Microscopy Sciences). The Gram reaction was determined using the KOH-based non-staining method (Powers, 1995). Catalase activity was tested by applying a 3% (v/v) hydrogen peroxide solution, and oxidase activity was assessed with 1% (w/v) Kovac’s reagent (bioMérieux). Bacterial motility was evaluated by stabbing into marine soft agar containing 0.5% (w/v) agar. Growth temperature range and optimum were determined on marine agar (MA) at 4°C, 10–30°C (at 5°C intervals), 37°C, and 42°C. Salt tolerance was tested on NaCl-free MA supplemented with NaCl at final concentrations ranging from 0% to 20% (w/v), in 0.5% increments from 0% to 4%, and at additional concentrations of 5%, 7.5%, 10%, 15%, and 20%. The pH range and optimum for growth were assessed in marine broth adjusted to pH 5.0–10.0 (in 1.0-unit intervals), using appropriate buffer systems: citrate (pH 4.0), MES (pH 5.0), MOPS (pH 6.0), HEPES (pH 7.0), and CHES (pH 8.0–11.0). Hydrolytic activities were evaluated on MA supplemented with the following substrates: starch (1%, w/v), colloidal chitin (1%, w/v), casein (3% skim milk, w/v), CM-cellulose (1%, w/v), Tween 20 (1%, v/v), and Tween 80 (1%, v/v). DNA degradation was tested on DNase test agar (BD Diagnostics) supplemented with 2% (w/v) NaCl. Hydrogen sulfide (H₂S) production was assessed using triple sugar iron (TSI) agar (BD Diagnostics) containing 2% (w/v) NaCl. Additional biochemical characteristics were determined using API 20NE and API ZYM strips (bioMérieux), following the manufacturer’s protocols with medium salinity adjusted to 2% (w/v) NaCl.
Fatty acid methyl ester (FAME) analysis was performed using biomass harvested from strains IMCC43871T, IMCC45206T, R. marina SAORIC-28T, and R. profundi SAORIC-476T grown on MA at 25°C for 7 d. FAME profiles were obtained by gas chromatography (Agilent 7890 GC) and interpreted using the Sherlock Microbial Identification System (MIDI), version 6.1, with the TSBA6 database (Sasser, 1990). Polar lipids were extracted according to the method of Minnikin et al. (1984) and separated by two-dimensional thin-layer chromatography (TLC) on silica gel 60 F254 plates. Total polar lipids were visualized using molybdatophosphoric acid, and functional group-specific detection was performed with ninhydrin (aminolipids), molybdenum blue (phospholipids), α-naphthol (glycolipids), and Dragendorff’s reagent (phosphatidylcholine). Respiratory quinones were analyzed by reverse-phase partition chromatography using Merck HPTLC RP-18 F254 plates following the method of Collins et al. (1980).
Nucleotide sequence accession numbers
The 16S rRNA gene sequences of strains IMCC43871T and IMCC45206T have been deposited in the GenBank/EMBL/DDBJ database under accession numbers PV186484 and PV076729, respectively. The complete genome sequences of IMCC43871T and IMCC45206T are available under accession numbers CP177355 and CP177357, respectively.
Results and Discussion
16S rRNA gene-based phylogeny
The 16S rRNA gene sequence similarity and phylogenetic analyses revealed that strains IMCC43871T and IMCC45206T are affiliated with the genus Rubrivirga. The two strains shared 99.2% 16S rRNA gene sequence similarity, which is slightly above the generally accepted species delineation threshold of 98.7% (Chun et al., 2018; Kim et al., 2014). Given this high level of 16S rRNA gene sequence similarity between the two strains, classification of the two strains as separate novel species required further genomic demarcation. Strains IMCC43871T and IMCC45206T showed the highest sequence similarity to R. marina SAORIC-28T (98.1% and 98.4%, respectively), followed by R. profundi SAORIC-476T (97.8% and 97.9%) and R. litoralis F349T (97.1% and 97.0%). Since the 16S rRNA gene sequence similarities between the two strains and the validly published Rubrivirga species were all below the 98.7% cutoff, the two strains were considered to represent independent species within the genus Rubrivirga. Phylogenetic analyses based on 16S rRNA gene sequences consistently demonstrated strains IMCC43871T and IMCC45206T formed a robust clade with the three Rubrivirga species, supported by high bootstrap values (Fig. 1), thereby confirming their taxonomic affiliation with the genus Rubrivirga. The congruence among treeing methods and consistently high bootstrap values across major nodes suggest that the phylogenetic positions of the novel strains are robustly resolved.
Phylogenomic analysis and genome characteristics
The complete genome sequence of strain IMCC43871T was assembled into two contigs comprising a circular chromosome and a 28,518 bp plasmid, whereas the genome of strain IMCC45206T was assembled into a single circular chromosome. The chromosomal genome sizes were 4,173,969 bp for strain IMCC43871T and 4,167,368 bp for strain IMCC45206T, with G + C contents of 71.8% and 72.8%, respectively. CheckM analysis estimated the genome completeness and contamination at 98.4% and 1.9% for both strains, respectively, indicating a high-quality genome assembly with over 95% completeness and less than 5% contamination.
A total of 3,620 and 3,593 protein-coding sequences, 58 and 54 tRNA genes, and three rRNA genes were identified in strains IMCC43871T and IMCC45206T, respectively. The complete 16 rRNA genes extracted from the genomes were identical (100%) to the amplified 16S rRNA gene sequences of the corresponding strains. An overview of the general genomic features of IMCC43871T, IMCC45206T, R. marina SAORIC-28T and R. profundi SAORIC-476T is provided in Table S1.
Genomic relatedness among IMCC43871T, IMCC45206T, and Rubrivirga species were assessed by calculating ANI and dDDH values. The ANI and dDDH values between IMCC43871T and IMCC45206T were 88.5% and 36.3%, respectively, below the established thresholds of 95−96% for ANI and 70% for dDDH proposed for bacterial species demarcation (Chun et al., 2018; Riesco and Trujillo, 2024), confirming that each strain represents a distinct novel species. Strain IMCC43871T exhibited ANI values of 77.5% with R. marina SAORIC-28T, 77.4% with R. profundi SAORIC-476T, and 77.2% with R. litoralis F349T, while the corresponding dDDH values were 20.8%, 21.4%, and 21.3%, respectively. Similarly, IMCC45206T showed ANI values of 77.7%, 77.4%, and 77.6%, and dDDH values of 21.3%, 21.4%, and 21.2% with R. marina SAORIC-28T, R. profundi SAORIC-476T, and R. litoralis F349T, respectively. All these values were well below the species delineation thresholds of ANI and dDDH, supporting the classification of strains IMCC43871T and IMCC45206T as two novel species within the genus. In the genome-based phylogenetic tree, strains IMCC43871T and IMCC45206T formed a distinct and well-supported clade within the genus Rubrivirga, further supporting their taxonomic assignment into the genus Rubrivirga (Fig. 2).
Functional annotation revealed that the genomes of IMCC43871T and IMCC45206T encode core metabolic pathways associated with central carbon metabolism, including the Entner–Doudoroff pathway, tricarboxylic acid cycle, pentose phosphate pathway, and phosphoribosyl pyrophosphate biosynthesis, showing the typical heterotrophic lifestyle. Both strains also harbor gene clusters related to flagellar biosynthesis; however, flagella were not observed under transmission electron microscopy. Strains IMCC43871T and IMCC45206T exhibited overall COG functional category distributions similar to those of other Rubrivirga species, but relatively higher proportions in categories such as coenzyme transport and metabolism, lipid transport and metabolism, and secondary metabolism may indicate adaptations to environmental fluctuations typical of coastal habitats (Table S2).
Physiological and chemotaxonomic characteristics
Transmission electron microscopy revealed that cells of strains IMCC43871T and IMCC45206T were irregular short rods, with approximate dimensions of 0.7–0.9 µm × 1.4–2.4 µm and 0.7–0.9 µm × 1.6–2.4 µm, respectively (Fig. S1). Although genome annotation indicated the presence of genes associated with flagellar biosynthesis in both strains, flagella were not observed in the TEM images. The physiological and biochemical characteristics of strains IMCC43871T and IMCC45206T, along with those of two closely related Rubrivirga species, are summarized in Table 1 and the species protologues. Strains IMCC43871T and IMCC45206T exhibited similar physiological traits, including optimal growth conditions, oxidase and catalase activities, and the ability to hydrolyze Tween 20 and Tween 80, but differed from each other in several enzyme activities. In addition, both strains could be distinguished from the closely related species of the genus Rubrivirga by differences in growth properties and certain enzymatic activities.
The fatty acid compositions of IMCC43871T, IMCC45206T, and two Rubrivirga species are presented in Table 2. The fatty acid profiles of the four strains were generally similar, with all strains containing C17 :1 ω8c and summed feature 9 (iso-C17:1 ω9c and/or C16:0 10-methyl) as predominant components, reflecting their affiliation within the same genus. Strains IMCC43871T and IMCC45206T shared major fatty acids present at > 10%, including C17 :1 ω8c (22.3% and 20.4%, respectively) and summed feature 9 (25.2% and 25.4%), indicating close relatedness between the two novel strains. However, the two novel strains exhibited notable differences from R. marina and R. profundi, particularly in the proportions of certain fatty acids such as iso-C17:0 and summed feature 3 (C16:1 ω7c and/or C16:1 ω6c). These compositional variations in branched-chain fatty acids and specific unsaturated fatty acids further differentiate the two novel strains from previously described Rubrivirga species.
The respiratory quinone detected in both novel strains was menaquinone-7 (MK-7). The major polar lipids of strain IMCC43871T included phosphatidylethanolamine (PE), phosphatidylglycerol (PG), diphosphatidylglycerol (DPG), three unidentified phospholipids, and four unidentified lipids (Fig. S2). Similarly, strain IMCC45206T contained PE, PG, DPG, two unidentified phospholipids, and five unidentified lipids. Both strains shared the presence of PE, PG, and DPG, consistent with the lipid profiles reported for other Rubrivirga species. Despite sharing PE, PG, and DPG as major polar lipids, the novel strains differed from previously described Rubrivirga species in possessing distinct sets and numbers of unidentified lipids. The fatty acid composition, respiratory quinone type, and polar lipid profiles of strains IMCC43871T and IMCC45206T support their chemotaxonomic placement within the genus Rubrivirga.
Taxonomic conclusion
Collective evidence from 16S rRNA gene- and genome-based phylogenetic analyses, along with chemotaxonomic characteristics, clearly indicated that strains IMCC43871T and IMCC45206T are assigned to the genus Rubrivirga. However, the low levels of genomic relatedness, as indicated by ANI and dDDH values well below the species delineation thresholds, together with distinct phenotypic and chemotaxonomic characteristics, clearly demonstrate that these two strains represent two novel and distinct species within the genus. Accordingly, the names Rubrivirga aquatilis sp. nov. and Rubrivirga halophila sp. nov. are proposed for strains IMCC43871T and IMCC45206T, respectively. Beyond their taxonomic novelty, the lipid-hydrolyzing activity of the two novel species suggests ecological relevance in organic matter degradation and potential utility in marine biotechnology.
Description of Rubrivirga aquatilis sp. nov.
Rubrivirga aquatilis (a.qua'ti.lis. L. adj. aquatic, pertaining to water, referring to the marine origin of the type strain).
Cells are Gram-stain-negative, obligately aerobic, non-motile rods, measuring approximately 0.7–0.9 µm in width and 1.4–2.4 µm in length. Colonies grown on marine agar are pink, circular, and convex. Growth occurs at 15–30°C (optimum, 25°C), pH 7.0–8.0 (optimum, pH 7.0), and in the presence of 1.0–4.0% (w/v) NaCl (optimum, 2.0%). Both oxidase and catalase activities are positive. In API 20NE tests, the strain is negative for nitrate reduction, esculin hydrolysis, β-galactosidase, indole production, glucose fermentation, arginine dihydrolase, urease, and gelatinase. In API ZYM tests, activities of alkaline phosphatase, esterase (C4), esterase lipase (C8), leucine arylamidase, valine arylamidase, trypsin, α-chymotrypsin, acid phosphatase, naphthol-AS-BI-phosphohydrolase, and N-acetyl-β-glucosaminidase are positive, whereas lipase (C14), cystine arylamidase, α-galactosidase, β-galactosidase, α-glucosidase, β-glucosidase, α-mannosidase, β-glucuronidase, and α-fucosidase are negative. Hydrolyzes Tween 20 and Tween 80, but does not hydrolyze casein, colloidal chitin, DNA, starch, or CM-cellulose. The predominant fatty acids are summed feature 9 (comprising iso-C17:1 ω9c and/or C16:0 10-methyl) and C17:1 ω8c. The major respiratory quinone is menaquinone-7 (MK-7). The polar lipid profile includes phosphatidylethanolamine, phosphatidylglycerol, diphosphatidylglycerol, three unidentified phospholipids, and four unidentified lipids. The type strain is IMCC43871T (= KCTC 102072T = NBRC 116463T = HNIBRBA6996T), isolated from coastal seawater in the Republic of Korea. The complete genome of the type strain is 4,173,969 bp in length, with a DNA G + C content of 71.8%. The GenBank accession numbers for the 16S rRNA gene sequence and genome sequence are PV186484 and CP177355, respectively.
Description of Rubrivirga halophila sp. nov.
Rubrivirga halophila (ha.lo'phi.la. Gr. hals, salt; Gr. philos, loving; N.L. fem. adj. halophila, salt-loving, referring to the marine, moderately halophilic nature of the species).
Cells are Gram-stain-negative, obligately aerobic, non-motile, short rod-shaped, measuring approximately 0.7–0.9 µm in width and 1.6–2.4 µm in length. Colonies formed on marine agar are pale pink, circular, soft, and convex. Growth occurs at 20–30°C (optimum, 25°C), at pH 7.0–8.0 (optimum, pH 7.0), and in the presence of 0.5–5.0% (w/v) NaCl (optimum, 2.0%). Both oxidase and catalase activities are positive. In API 20NE tests, esculin hydrolysis, gelatinase, and β-galactosidase are positive, whereas nitrate reduction, indole production, glucose fermentation, arginine dihydrolase, and urease are negative. In API ZYM tests, activities of alkaline phosphatase, esterase (C4), esterase lipase (C8), leucine arylamidase, valine arylamidase, α-chymotrypsin, acid phosphatase, naphthol-AS-BI-phosphohydrolase, and N-acetyl-β-glucosaminidase are positive. No activity is detected for lipase (C14), cystine arylamidase, trypsin, α-galactosidase, β-galactosidase, α-glucosidase, β-glucosidase, α-mannosidase, β-glucuronidase, or α-fucosidase. Hydrolyzes Tween 20 and Tween 80, but does not hydrolyze casein, colloidal chitin, DNA, starch, or CM-cellulose. The predominant fatty acids are summed feature 9 (comprising iso-C17:1 ω9c and/or C16:0 10-methyl) and C17:1 ω8c. The major respiratory quinone is menaquinone-7 (MK-7). The polar lipid profile includes phosphatidylethanolamine, phosphatidylglycerol, diphosphatidylglycerol, two unidentified phospholipids, and five unidentified lipids. The type strain is IMCC45206T (= KCTC 92925T = NBRC 116172T = CCTCC AB 2023136T = HNIBRBA14924T), isolated from coastal seawater in the Republic of Korea. The complete genome of the type strain is 4,167,368 bp in length, with a DNA G + C content of 72.8%. The GenBank accession numbers for the 16S rRNA gene and the genome sequence are PV076729 and CP177357, respectively.
Acknowledgments
This study was supported by the research grant “Survey of island-coastal indigenous organisms (Prokaryotes)” (HNIBR202301211; HNIBR202501213) from Honam National Institute of Biological Resources (HNIBR) in Korea and by the Mid-Career Research Program (NRF-2022R1A2C3008502) through the National Research Foundation (NRF) funded by the Ministry of Sciences and ICT.
Conflict of Interest
The authors declare no conflict of interest.
Supplementary Information
The online version contains supplementary material available at https://doi.org/10.71150/jm.2504017.
Table S1.
Comparative genomic features of strains IMCC43871T, IMCC45206T, Rubrivirga marina SAORIC-28T, and Rubrivirga profundi SAORIC-476T
Strains: 1, IMCC43871T; 2, IMCC45206T; 3, R. marina SAORIC-28T; 4, R. profundi SAORIC-476T
jm-2504017-Supplementary-Table-S1.pdf
Table S2.
Distribution of genes in general COG categories in the genomes of strains IMCC43871T, IMCC45206T, R. marina SAORIC-28T, and R. profundi SAORIC-476T
Strains: 1, IMCC43871T; 2, IMCC45206T; 3, R. marina SAORIC-28T; 4, R. profundi SAORIC-476T
jm-2504017-Supplementary-Table-S2.pdf
Fig. S1.
Transmission electron micrographs of cells of strains IMCC43871T (A), IMCC45206T (B) after negative staining with 0.5% (w/v) uranyl acetate. Cells were grown on marine agar at 25°C for 7 days. Bar, 0.2 μm.
jm-2504017-Supplementary-Fig-S1.pdf
Fig. S2.
Two-dimensional thin layer chromatograph of polar lipids of strains IMCC43871T (A) and IMCC45206T (B). Total lipids were detected by spraying with 5% molybdatophosphoric acid solution. PE, phosphatidylethanolamine; PG, phosphatidylglycerol; DPG, diphosphatidylglycerol, PL1-3, unidentified phospholipid; L1-5, unidentified lipid. 1, first dimension of TLC; 2, second dimension of TLC.
jm-2504017-Supplementary-Fig-S2.pdf
Fig. 1.Maximum-likelihood phylogenetic tree based on 16S rRNA gene sequences showing the positions of strains IMCC43871T and IMCC45206T. Bootstrap values (expressed as percentages of 1,000 replications) over 75% are shown at nodes for maximum-likelihood, neighbor-joining, and minimum-evolution methods, respectively. Filled circles indicate that the corresponding nodes were recovered by all treeing methods. Open circles indicate that the corresponding nodes were recovered by any two out of three. Bar, 0.05 substitutions per nucleotide position.
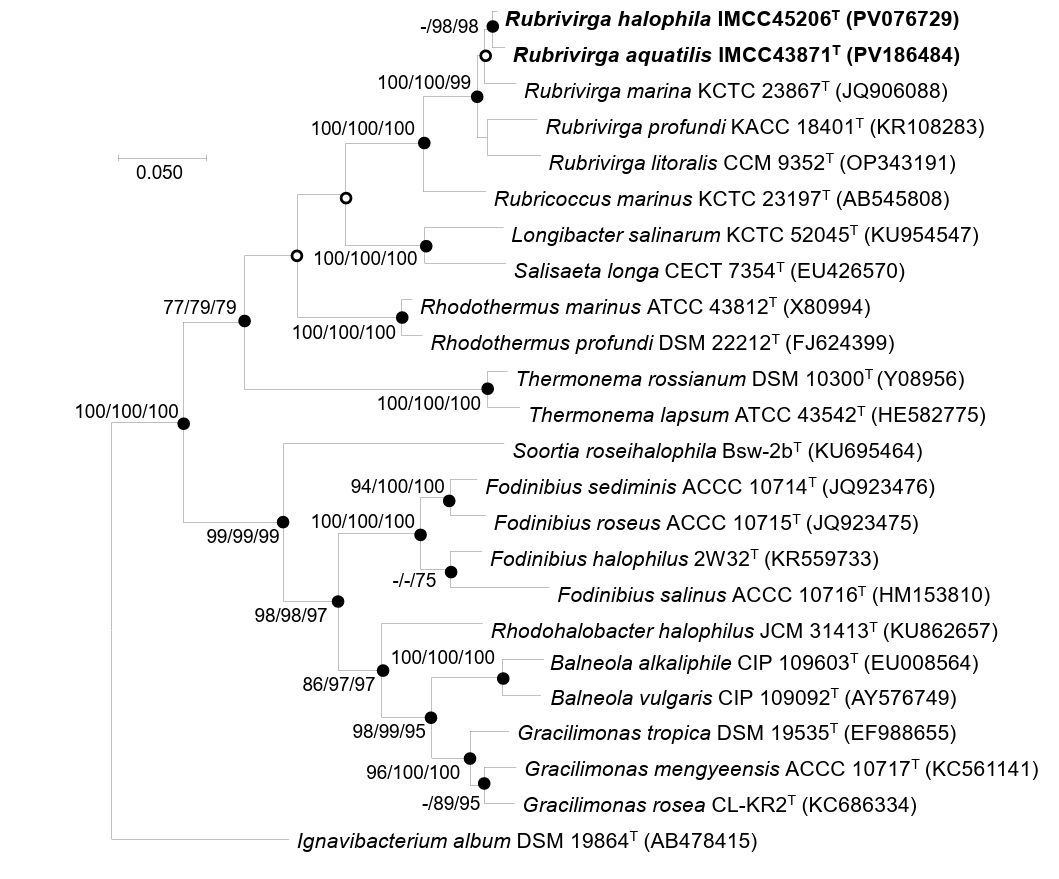
Fig. 2.Phylogenomic tree based on concatenated multiple alignments of 81 genes showing the positions of strains IMCC43871T and IMCC45206T. Calculations of bootstrap values are shown at nodes. Bar, 0.2 substitutions per amino acid position.

Table 1.
Differential phenotypic characteristics of strains IMCC43871T and IMCC45206T and closely related type strains of the genus Rubrivirga.
Strains: 1, IMCC43871T; 2, IMCC45206T; 3, R. marina SAORIC-28T; 4, R. profundi SAORIC-476T. Data were obtained from this study. All strains were cultured under identical conditions. All strains are positive for oxidase, catalase, alkaline phosphatase, esterase (C4), esterase lipase (C8), leucine arylamidase, valine arylamidase, acid phosphatase, naphthol-AS-BI-phosphohydrolase, and N-acetyl-β-glucosaminidase. +, positive; ‒, negative
|
Characteristics |
1 |
2 |
3 |
4 |
|
Cell size (µm) |
0.7–0.9 × 1.4–2.4 |
0.7–0.9 × 1.6–2.4 |
0.2–1.2 × 0.7–5.3 |
0.6–0.8 × 1.8–3.0 |
|
Colony color |
pale pink |
pink |
pale red |
pale red |
|
Growth at/in |
|
|
|
|
|
Optimum temperature (℃) |
25 |
25 |
25–30 |
25 |
|
pH range |
7.0–8.0 |
7.0–8.0 |
6.0–9.0 |
6.0–8.5 |
|
Optimum pH |
7 |
7 |
6.0–8.0 |
7.5 |
|
NaCl range (%, w/v) |
1–4 |
0.5–5 |
1–5 |
1–5 |
|
API 20NE |
|
|
|
|
|
Esculin hydrolysis |
‒ |
+ |
+ |
+ |
|
Gelatinase |
‒ |
+ |
+ |
+ |
|
β-Galactosidase (PNPG) |
‒ |
+ |
+ |
+ |
|
API ZYM |
|
|
|
|
|
Crystine arylamidase |
‒ |
‒ |
‒ |
+ |
|
Trypsin |
+ |
‒ |
‒ |
‒ |
|
β-Glucuronidase |
‒ |
‒ |
‒ |
+ |
|
α-Chymotrypsin |
+ |
+ |
‒ |
‒ |
Table 2.
Cellular fatty acid compositions of strains IMCC43871T and IMCC45206T and the closely related type strains of the genus Rubrivirga.
Strains: 1, IMCC43871T; 2, IMCC45206T; 3, R. marina SAORIC-28T; 4, R. profundi SAORIC-476T. Data were obtained from this study. All strains were cultured under identical conditions. Fatty acids comprising < 1.0% of the total fatty acid content in all species were omitted. ‒, Not detected; Tr, traces (< 1.0%). Major fatty acids (> 10%) are shown in bold.
|
Fatty acid (%) |
1 |
2 |
3 |
4 |
|
Saturated
|
|
|
|
|
|
C16:0
|
5.5 |
4.5 |
3.8 |
5.6 |
|
C17:0
|
4.2 |
7.9 |
3.9 |
4.7 |
|
Branched
|
|
|
|
|
|
iso-C14:0
|
– |
– |
1.5 |
– |
|
iso-C15:0
|
5.5 |
3.2 |
6.3 |
12.0
|
|
iso-C16:0
|
1.8 |
1.7 |
1.5 |
1.9 |
|
iso-C17:0
|
6.0 |
9.6 |
16.1
|
15.1
|
|
anteiso-C15:0
|
Tr |
Tr |
1.9 |
– |
|
anteiso-C17:0
|
1.1 |
Tr |
2.1 |
– |
|
Unsaturated
|
|
|
|
|
|
C15:1 ω6c |
Tr |
Tr |
1.3 |
3.8 |
|
C16:1 ω9c |
5.0 |
2.5 |
4.3 |
4.4 |
|
C15:1 ω8c |
2.2 |
1.1 |
– |
– |
|
C17:1 ω6c |
3.2 |
3.8 |
6.7 |
7.2 |
|
C17:1 ω8c |
22.3
|
20.4
|
12.6
|
12.7
|
|
C18:1 ω9c |
3.9 |
3.2 |
4.7 |
– |
|
iso-C15:1 F |
1.1 |
Tr |
– |
– |
|
Summed feature*
|
|
|
|
|
|
3 |
7.6 |
4.6 |
13.6
|
8.7 |
|
9 |
25.2
|
25.4
|
20.0
|
23.8
|
References
- Bouras G, Houtak G, Wick RR, Mallawaarachchi V, Roach MJ, et al. 2024. Hybracter: enabling scalable, automated, complete and accurate bacterial genome assemblies. Microb Genom. 10: 001244.ArticlePubMedPMC
- Chun J, Oren A, Ventosa A, Christensen H, Arahal DR, et al. 2018. Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int J Syst Evol Microbiol. 68: 461–466. ArticlePubMed
- Collins MD, Shah HN, Minnikin DE. 1980. A note on the separation of natural mixtures of bacterial menaquinones using reverse phase thin-layer chromatography. J Appl Bacteriol. 48: 277–282. ArticlePubMed
- Felsenstein J. 1981. Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol. 17: 368–376. ArticlePubMedPDF
- Felsenstein J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 39: 783–791. ArticlePubMedPDF
- Fitch WM. 1971. Toward defining the course of evolution: minimum change for a specific tree topology. Systematic Zoology. 20: 406–416. Article
- Han D, Shin H, Lee JH, Kang CK, Kim DG, et al. 2022. Phylogenetic diversity and spatiotemporal dynamics of bacterial and microeukaryotic plankton communities in Gwangyang Bay of the Korean Peninsula. Sci Rep. 12: 2980.ArticlePubMedPMCPDF
- Kanehisa M, Sato Y, Morishima K. 2016. BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J Mol Biol. 428: 726–731. ArticlePubMed
- Kim J, Na SI, Kim D, Chun J. 2021. UBCG2: Up-to-date bacterial core genes and pipeline for phylogenomic analysis. J Microbiol. 59: 609–615. ArticlePubMedPDF
- Kim M, Oh HS, Park SC, Chun J. 2014. Towards a taxonomic coherence between average nucleotide identity and 16s rRNA gene sequence similarity for species demarcation of prokaryotes. Int J Syst Evol Microbiol. 64: 346–351. ArticlePubMed
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35: 1547–1549. ArticlePubMedPMC
- Lee J, Song SH, Moon K, Lee N, Ryu S, et al. 2024. Thalassotalea aquiviva sp. nov., and Thalassotalea maritima sp. nov., isolated from Seawater of the Coast in South Korea. J Microbiol. 62: 1099–1111. ArticlePubMedPDF
- Ludwig W, Strunk O, Westram R, Richter L, Meier H, et al. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32: 1363–1371. ArticlePubMedPMC
- Marchler-Bauer A, Zheng C, Chitsaz F, Derbyshire MK, Geer LY, et al. 2013. CDD: conserved domains and protein three-dimensional structure. Nucleic Acids Res. 41: D348–D352. ArticlePubMed
- Meier-Kolthoff JP, Auch AF, Klenk HP, Goker M. 2013. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics. 14: 60.ArticlePubMedPMCPDF
- Minnikin DE, O'Donnell AG, Goodfellow M, Alderson G, Athalye M, et al. 1984. An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J Microbiol Methods. 2: 233–241. Article
- Park S, Song J, Yoshizawa S, Choi A, Cho JC, et al. 2013. Rubrivirga marina gen. nov., sp. nov., a member of the family rhodothermaceae isolated from deep seawater. Int J Syst Evol Microbiol. 63: 2229–2233. ArticlePubMed
- Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. 2015. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 25: 1043–1055. ArticlePubMedPMC
- Powers EM. 1995. Efficacy of the ryu nonstaining KOH technique for rapidly determining gram reactions of food-borne and waterborne bacteria and yeasts. Appl Environ Microbiol. 61: 3756–3758. ArticlePubMedPMCPDF
- Rey-Velasco X, Lucena T, Belda A, Gasol JM, Sanchez O, et al. 2024. Genomic and phenotypic characterization of 26 novel marine bacterial strains with relevant biogeochemical roles and widespread presence across the global ocean. Front Microbiol. 15: 1407904.ArticlePubMedPMC
- Richter M, Rosselló-Móra R, Oliver Glöckner F, Peplies J. 2016. JSpeciesWS: a web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics. 32: 929–931. ArticlePubMedPDF
- Riesco R, Trujillo ME. 2024. Update on the proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int J Syst Evol Microbiol. 74: 006300.ArticlePubMedPMC
- Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 4: 406–425. ArticlePubMed
- Sasser M. 1990. Identification of bacteria by gas chromatography of cellular fatty acids. Technical Note #101:1–7. MIDI Inc..
- Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 30: 2068–2069. ArticlePubMedPDF
- Song J, Joung Y, Park S, Cho JC, Kogure K. 2016. Rubrivirga profundi sp. nov., isolated from deep-sea water, and emended description of the genus Rubrivirga. Int J Syst Evol Microbiol. 66: 3253–3257. ArticlePubMed
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30: 1312–1313. ArticlePubMedPMCPDF
- Tak H, Park MS, Cho H, Lim Y, Cho JC. 2024. Congregibacter variabilis sp. nov. and Congregibacter brevis sp. nov. within the OM60/NOR5 clade, isolated from seawater, and emended description of the genus Congregibacter. J Microbiol. 62: 739–748. ArticlePubMedPDF
- Wainwright BJ, Millar T, Bowen L, Semon L, Hickman KJE, et al. 2023. The core mangrove microbiome reveals shared taxa potentially involved in nutrient cycling and promoting host survival. Environ Microbiome. 18: 47.ArticlePubMedPMCPDF
- Weisburg WG, Barns SM, Pelletier DA, Lane DJ. 1991. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol. 173: 697–703. ArticlePubMedPMCPDF
- Yang SH, Park MJ, Oh HM, Kwon KK. 2023. Description of Fervidibacillus gen. nov. with two species, Fervidibacillus albus sp. nov. and Fervidibacillus halotolerans sp. nov., isolated from tidal flat sediments, and emendation of misclassified taxa in the genus Caldibacillus. J Microbiol. 61: 175–187. ArticlePubMedPDF
- Yang SH, Park MJ, Oh HM, Park YJ, Kwon KK. 2024. Flavivirga spongiicola sp. nov. and Flavivirga abyssicola sp. nov., isolated from marine environments. J Microbiol. 62: 11–19. ArticlePubMedPDF
- Yoon SH, Ha SM, Kwon S, Lim J, Kim Y, et al. 2017. Introducing EzBioCloud: A taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int J Syst Evol Microbiol. 67: 1613–1617. ArticlePubMedPMC
- Zheng Y, Wang Y, Dan M, Li Y, Zhao G, et al. 2023. Characterization of degradation patterns and enzymatic properties of a novel alkali-resistant alginate lyase AlyRm1 from Rubrivirga marina. Curr Res Food Sci. 6: 100414.ArticlePubMed
Citations
Citations to this article as recorded by




 ePub Link
ePub Link Cite this Article
Cite this Article

 MSK
MSK





