ABSTRACT
- Phage specificity primarily relies on host cell-surface receptors. However, integrating cas genes and guide RNAs into phage genomes could enhance their target specificity and regulatory effects. In this study, we developed a CRISPR-Cas12f1 system-equipped bacteriophage λ model capable of detecting Escherichia coli target genes. We demonstrated that synthetic λ phages carrying Cas12f1-sgRNA can effectively prevent lysogen formation. Furthermore, we showcased that truncating the 3'-end of sgRNA enables precise identification of single-nucleotide variations in the host genome. Moreover, infecting E. coli strains carrying various stx2 gene subtypes encoding Shiga toxin with bacteriophages harboring Cas12f1 and truncated sgRNAs resulted in the targeted elimination of strains with matching subtype genes. These findings underscore the ability of phages equipped with the CRISPR-Cas12f1 system to precisely control microbial hosts by recognizing genomic sequences with high resolution.
-
Keywords: bacteriophage, CRISPR-Cas, single base mismatch, bacterial control
Introduction
Bacteriophages are crucial for microbial regulation, offering alternatives to antibiotics due to their ability to kill host microbes (Rogovski et al., 2021). However, narrow host ranges and lysogeny formation can limit their efficacy (Lin et al., 2022). Nevertheless, advances in phage engineering have enabled the creation of synthetic phages with enhanced applications for microbial regulation (Jia et al., 2023; Lobocka et al., 2021). For instance, synthetic phages with altered host ranges can be produced using cell-free in vitro phage rebooting technology (Liang et al., 2022), and mutagenesis of phage tail fibers can modify host specificity (Yehl et al., 2019).
CRISPR-Cas systems are pivotal for genome editing in bacteriophages because they facilitate easy alteration of DNA specificity through guide RNA modules (Sternberg and Doudna, 2015). Duong et al. (2020) engineered a light-emitting synthetic phage by inserting the luciferase-encoding gene into the outer capsid protein gene of T4 phage using CRISPR-Cas9. Guan et al. (2022) used CRISPR-Cas13a to label the coat protein of the ФKZ jumbo phage with a fluorescent tag. Furthermore, CRISPR-Cas9 has been employed to invert the direction of the repressor cI gene in the temperate phage λ, converting it into an obligate lytic phage (Lee et al., 2022).
Additionally, phages can be used to deliver CRISPR-Cas systems to host bacteria. Nethery et al. (2022) developed a system that specifically edits Escherichia coli in a simulated co-culture of three microbial species using a cytosine base editor-equipped λ phage. Yosef et al. (2015) developed a system in which a Type I-E CRISPR-loaded λ synthetic phage removes antibiotic resistance plasmids from lysogenic strains, rendering pathogens susceptible to antibiotics.
Recently, the CRISPR-Cas system has been repurposed to specifically control target microbes. For instance, cas9-sgRNA genes were transferred from E. coli to Salmonella enterica through conjugation, resulting in the selective killing of S. enterica (Hamilton et al., 2019). M13-phagemids carrying the cas9 gene (Lam et al., 2021) or non-replicating phages with inserted cas9 genes (Mitsunaka et al., 2022) were engineered to specifically kill E. coli strains harboring target genes in the genome. Selle et al. (2020) reported that synthetic phages, with CRISPR arrays incorporated into the genome of ϕCD24-2 phages and key lysogeny genes removed, specifically killed Clostridium difficile and inhibited lysogeny in mouse colonic tissues.
Efforts have been made to enhance regulatory effects by incorporating cas genes and guide RNAs into phage genomes. Park et al. (Park et al., 2017; Cobb et al., 2019) reported that integrating the cas9 gene into the genome of the temperate φ SaBov phage enhanced the killing effect of Staphylococcus aureus in mouse skin and rat osteomyelitis models. Jin et al. (2022) demonstrated that incorporating a 9.5 kb CRISPR-Cas3 cascade system into the λ genome would enable the specific control of enterohemorrhagic E. coli (EHEC) in an EHEC-infected mouse model. Gencay et al. (2024) incorporated a Type I-E CRISPR-Cas system into phage genomes to achieve phage-mediated specific killing of E. coli in the mouse gut environment. Previous studies have focused on regulating host bacteria by targeting specific genes. However, these methods often struggle to selectively control harmful bacterial strains that arise due to nucleotide variations, potentially affecting innocent cells.
In this study, we explored the potential for precisely controlling and suppressing lysogenic strain emergence by integrating the Cas12f1 system into the λ phage genome. With its compact size—approximately one-third that of commonly used Cas enzymes like Cas9 and Cas12a—Cas12f1 offers a significant advantage in facilitating seamless genomic incorporation (Kim et al., 2021b). Leveraging the single-base resolution of truncated guide RNAs obtained from previous studies (Lee et al., 2023), we aimed to identify and control target host bacteria based on single-base variations and genotypes in pathogenic microbe-like strain models using Cas12f1-loaded phages. Finally, based on the experimental results, we explored the prospects of precise microbial genome recognition and discussed control methods utilizing CRISPR-loaded phages.
Materials and Methods
Strains and cultivation
The E. coli strains used in this study are listed in Table S1. These strains were cultured in LB broth at 30°C or 37°C, depending on the specific requirements of each strain. To produce competent cells, the E. coli strains were cultured overnight at 30°C and then inoculated into LB broth at a final volume of 1%. The cultures were maintained at 30°C until the OD600 reached 0.4, at which point they were harvested. Strains MG1655 and HL051 harboring the pKD46 plasmid with λ red recombinase were further cultured for 3 h with a 1 mM final concentration of L-arabinose. Subsequently, the cultures were washed twice with 10% glycerol and aliquoted into 50 μl volumes for storage at -80°C. Depending on the selection marker of the plasmid or genetic cassette, ampicillin, kanamycin, chloramphenicol, and spectinomycin were added to the media at final concentrations of 50, 25, 12.5, and 75 μg/ml, respectively.
Lysogeny construction
The HL051 strain (λ-lysogenic MG1655) was generated as follows: 150 μl of an MG1655 overnight culture was added to 15 ml of 0.6% soft agar supplemented with CaCl2 (5 mM) and MgSO4 (10 mM), mixed, and overlaid onto an LB agar plate (90 mm diameter). After air-drying for 20 min, 4 μl of λ cI857 phage lysate was spotted on the agar, and the plate was incubated at 24°C for a week. Colonies from turbid spots were isolated, and lysogenic strain formation was confirmed using PCR with the primer pair (attB_F + λint_R) targeting the bacterial and phage genomes. Oligonucleotides for PCR are listed in Table S2.
Host engineering
E. coli HL059 (MG1655 galK 504AT) was edited using CRISPR-Cas9, and then the cas9 gene from the arabinose operon was removed via P1 transduction and the original araBAD operon was reintroduced. E. coli HL066 (MG1655 ΔgalK) was generated by introducing the ΔgalK mutation from the Keio collection JW0740 strain into MG1655 through P1 transduction (Baba et al., 2006). To produce an MG1655 strain carrying the stx2 gene, the stx2a gene was amplified using genomic DNA from ATCC 43895 as a template, and the stx2a-CmR cassette was constructed and inserted by replacing the srlAEBD operon in the MG1655 genome. The stx2a-CmR cassette was inserted into the L-arabinose-induced MG1655 cells harboring the pKD46 plasmid through electroporation using a 0.1 cm cuvette at 25 µF, 200 Ω, and 1.8 kV. All subsequent electroporations were performed under the same conditions. After electroporation, 950 µl of the SOC medium was added to the cells, which were then allowed to recover at 30°C for 1 h and subsequently spread on chloramphenicol-containing LB agar and incubated at 37°C. The grown colonies were confirmed using PCR and further verified through Sanger sequencing to ensure that the generated HL085 strain carried the stx2a-CmR cassette. Nucleotide sequences for stx2 subtypes were referenced from the following NCBI accession numbers: stx2a (CP008957) and stx2g (AY286000). Genomic DNA from the HL085 strain was used as a template for overlap PCR to amplify the stx2g-CmR cassette, and HL086 (MG1655-stx2g) was produced in the same manner as the HL085 (MG1655-stx2a) strain.
cas12f1 integration
To insert the cas12f1 gene into the b2 region of the λ phage genome from which the genes ea59, ea47, and ea31 were deleted, a cas12f1-CmR cassette was constructed. The PrpsL-cas12f1 fragment was generated using p15a-AsCas12f-apmR (a gift from Quanjiang Ji; Addgene plasmid #171610) as a template. The CmR cassette was amplified using genomic DNA from HK1020 as a template (Kim et al., 2021a). An overlapping PCR was performed to create the PrpsL-cas12f1-CmR cassette, which was then electroporated into L-arabinose-induced HL051 (λ cI857 lysogen) cells harboring the pKD46 plasmid. The electroporated cells were spread on chloramphenicol-containing LB agar plates and incubated at 30°C. The resulting colonies were initially verified using PCR and further confirmed through Sanger sequencing to ensure the insertion of the cas12f1-CmR cassette into the b2 region of the λ prophage genome in the finally constructed HL062 strain (λ cI857 lysogen carrying cas12f1 gene).
cas12f1-sgRNA integration
Using the pHL267 plasmid (Lee et al., 2023) as a template, the sgRNA gene was amplified with the primer pair sgRNA_sacI_2F_N + KmR_sgRNA_N2_R. A 667 bp fragment of the cas12f1 gene was amplified using genomic DNA from the HL061 strain as a template (Lee et al., 2023). The KmR cassette was amplified using genomic DNA from the HL002 strain and the primer pair sgRNA_KmR_N2_F + b2_KmR_int_R. An overlapping PCR was performed to construct a cas12f1 (667 bp)-sgRNA-KmR cassette, which was then electroporated into the HL080 strain (λ cI857 lysogen carrying the cas12f1 gene and ΔgalK mutation). The electroporated cells were spread on a kanamycin-containing LB agar plate and incubated at 30°C. Colony PCR and Sanger sequencing were performed to confirm the construction of the HL081 strain (λ cI857 lysogen carrying the cas12f1 gene and the sgRNA targeting galK). Subsequently, synthetic phages with a modified sgRNA target recognition sequence (TRS) within the prophage genome were constructed using overlapping PCR with the HL081 genome as a template.
Spotting assay
Various phage-lysogenic cells were cultured overnight at 30°C. The supernatant from these cultures was spread on soft agar to yield phage plaques. An MG1655 overnight culture was inoculated into LB medium supplemented with CaCl2 (5 mM) and MgSO4 (10 mM) and cultured at 30°C with shaking at 180 rpm until the OD600 reached 0.4. Subsequently, phage plaques were added to these bacterial cultures, which were then incubated at 42°C until complete lysis occurred. The supernatant obtained after centrifugation (3,000 rpm, 4°C, 30 min), containing the phage lysates, was used to quantify plaque-forming units (pfu) and then stored at 4°C after adding 0.1% of CHCl3. To confirm the infectivity of the engineered phages and their ability to form clear or turbid spots, a spotting assay was performed. Specifically, 150 µl of each host strain’s overnight culture was added to 15 ml of 0.6% soft agar supplemented with CaCl2 (5 mM) and MgSO4 (10 mM), mixed, and overlaid on an LB agar plate (90 mm diameter). Then, 4 µl of phage lysates (~109 pfu/ml) were spotted on the soft agar plates. The plates were incubated for 24 h at 30°C and 16 h at 37°C, respectively, depending on the specific requirements of each strain.
Broth culture
To measure growth, single colonies of each host strain were grown overnight as starter cultures in LB broth at 30°C or 37°C with shaking at 180 rpm. For the main culture, 200 ml of LB broth in a 1 L flask was inoculated with the overnight culture to a final concentration of 1% and then incubated at 30°C or 37°C at 180 rpm until the OD600 reached 0.4. Subsequently, 49 ml of this culture was transferred into each 125 ml disposable flask. To achieve a multiplicity of infection (MOI) of 0.1, 1 ml of phage lysate was diluted and added to each flask, which was then incubated at 30°C or 37°C. The OD600 was measured using a spectrophotometer (Biochrom Libra S70, Harvard Bioscience, Inc., USA). Nineteen hours after phage addition, each culture was plated on LB agar plates and incubated at 30°C for 18 h. The resulting colonies were streaked onto kanamycin-containing LB plates to obtain single colonies. The presence of lysogeny in the obtained single colonies was confirmed using PCR with the primer pairs attB_F + λint_R and attB_F + bioB_R.
Results
Lysogenization of bacteriophage λ in E. coli
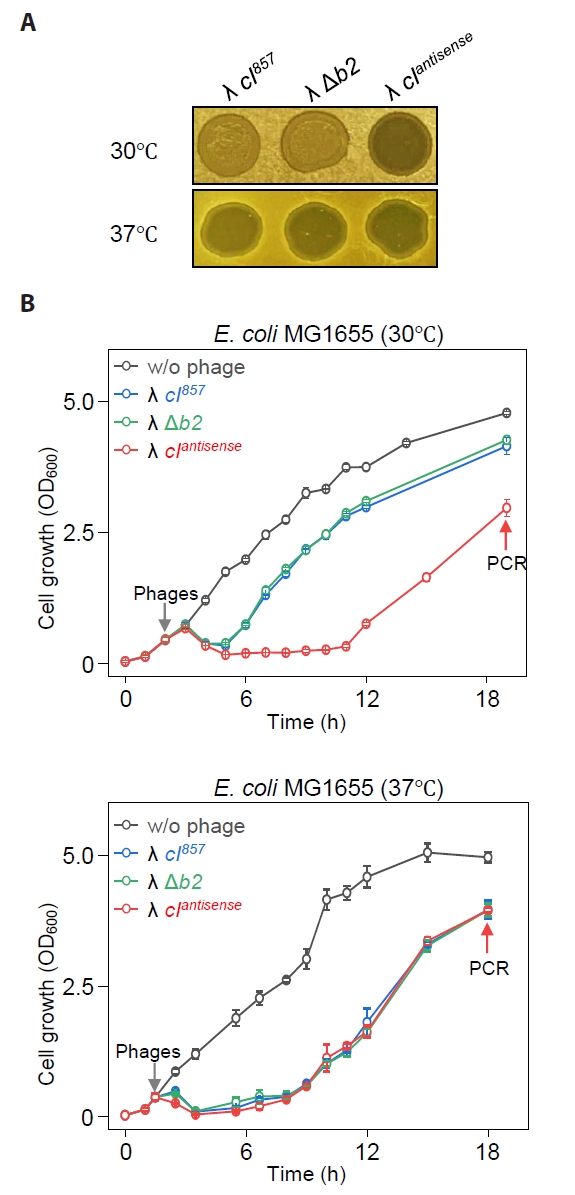
We compared the turbidity of spots formed on soft agar plates inoculated with host MG1655 and various phages: λ cI857 (George et al., 1987), λ Δb2 (with the non-essential gene removed), and the synthetic lytic phage λ cIantisense (Lee et al., 2022). The infection of λ cI857 and λ Δb2 predominantly follows the lysogenic cycle at 30°C but shifts to the lytic cycle at 37°C. In contrast, λ cIantisense exclusively undergoes the lytic cycle, regardless of temperature conditions. The results showed that λ cI857 formed turbid spots at 30°C and clear spots at 37°C. This indicates that at 30°C, lysogeny occurs, but at 37°C, the heat-labile nature of the CI857 repressor leads to its inactivation, triggering the λ lytic cycle (Fig. 1A). In contrast, the synthetic lytic phage λ cIantisense formed clear spots at all temperatures, as expected. Furthermore, λ Δb2 showed results similar to λ cI857, forming turbid spots at 30°C and clear spots at 37°C, suggesting that the removal of the b2 region does not affect the phage life cycle.
Subsequently, we infected liquid cultures of host MG1655 with these phages at 30°C and 37°C and measured growth to compare with the results observed in solid media. When MG1655 was infected with the lytic phage λ cIantisense at 30°C, complete lysis occurred by 3 h, and growth resumed at the 12 h point on the graph, reaching an OD600 value of 3.0 by 19 h (Fig. 1B). Infections with λ cI857 and λ Δb2 resulted in cell lysis, but cells began to regrow at 4 h post-infection, reaching an OD600 value of 4.2 by 19 h. Cells that regrew at the 19 h point (Fig. 1B) were no longer infected with λ cI857, and PCR results confirmed that the cells surviving λ cIantisense infection were phage-insensitive mutants. However, cells infected with λ cI857 and λ Δb2 were confirmed to be lysogenic (Fig. S1). When MG1655 was cultured at 37°C and separately infected with λ cI857, λ Δb2, and λ cIantisense, the patterns of host regrowth after lysis were the same. At 37°C, none of the regrown cells were infected with λ cI857, and PCR results confirmed that they were phage-insensitive mutant cells (Fig. S1).
These results demonstrate that λ cI857forms lysogens at 30°C but fails to do so at 37°C under both solid and liquid culture conditions. Additionally, the removal of the non-essential b2 region in λ Δb2 does not substantially impact the ability of the phage to infect hosts and form lysogens at 30°C.
Complete lysis of E. coli by bacteriophage λ with Cas12f1 nuclease
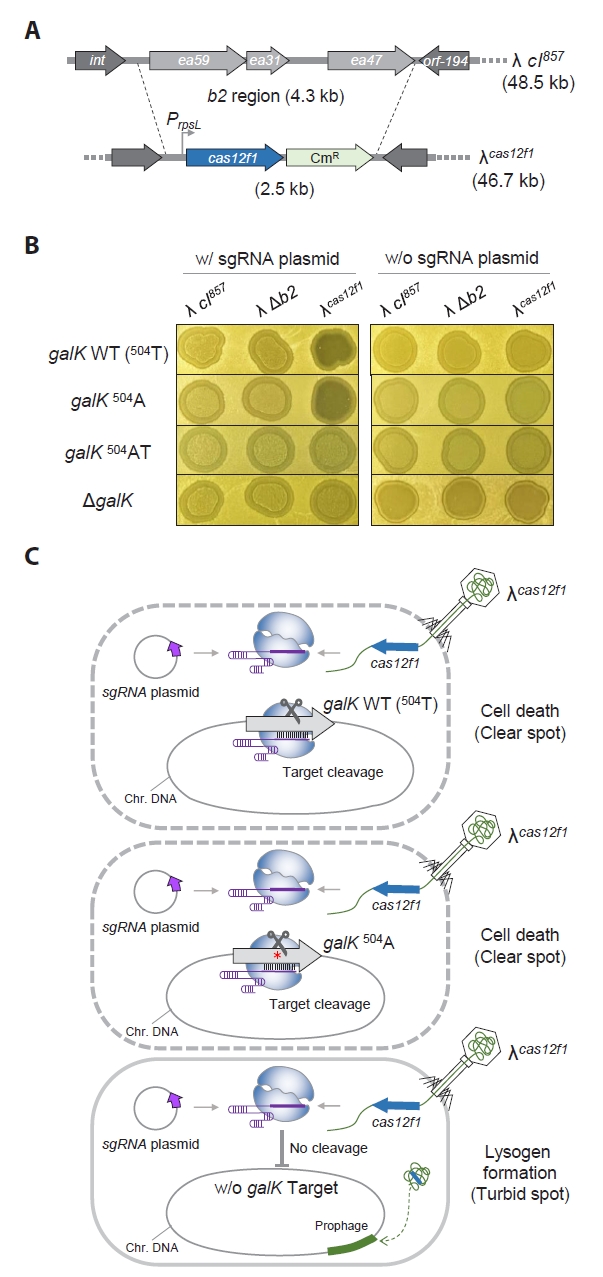
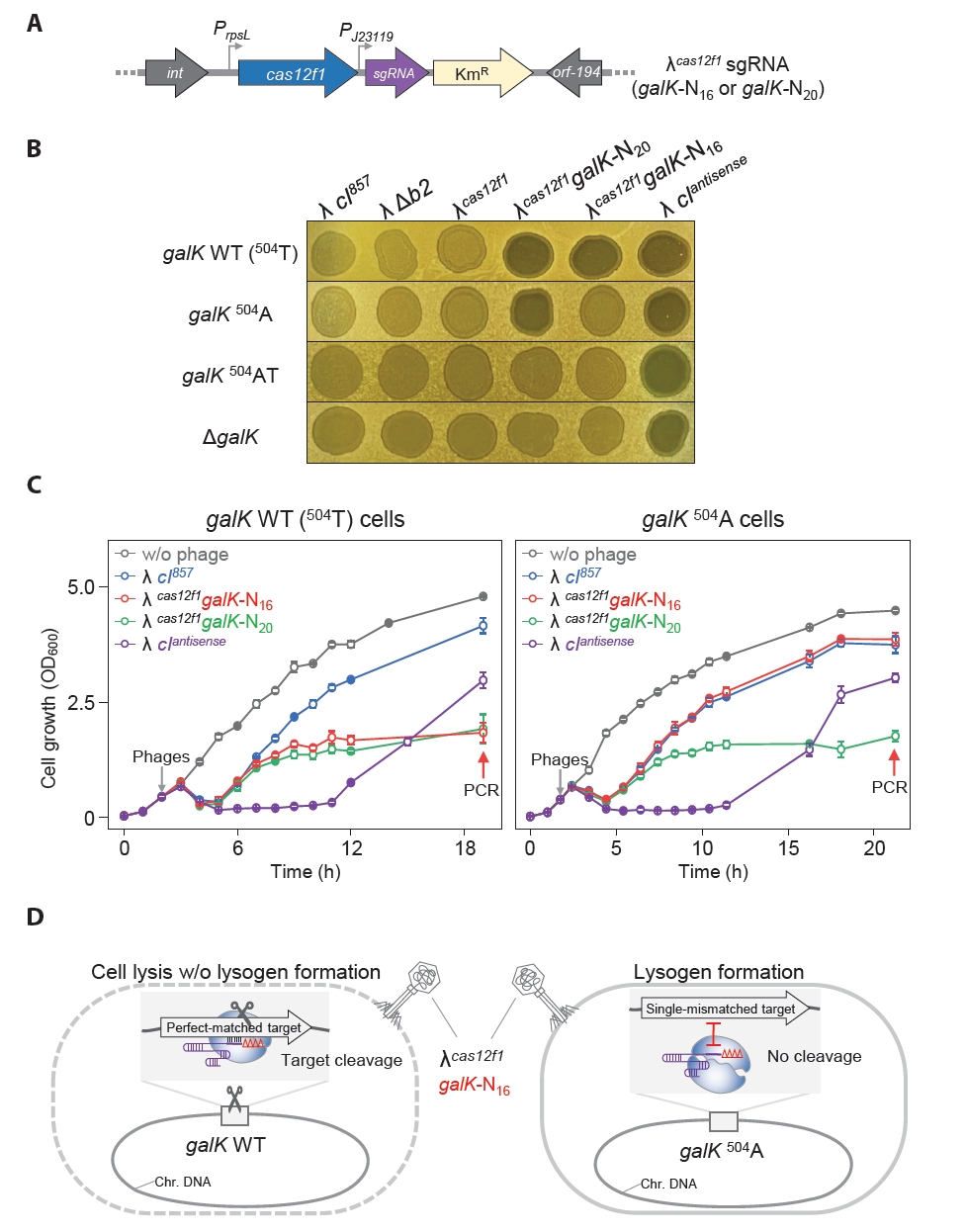
We constructed the λcas12f1 phage by inserting the cas12f1 gene at the location from which the b2 region was removed from the genome (Fig. 2A). To test the functionality of this engineered phage, we engineered E. coli cells with various galK target sequences—galK 504A, galK 504AT, and ΔgalK (Fig. S2)—and observed the turbidity of spots formed by the λcas12f1 phage on soft agar at 30°C, depending on the presence or absence of a galK-targeting sgRNA plasmid.
In cells harboring the sgRNA plasmid, clear spots were formed when the λcas12f1 phage was spotted on galK WT and galK 504A cells, while turbid spots were formed on galK 504AT and ΔgalK cells (Fig. 2B). All cells without the sgRNA plasmid formed turbid spots. These results indicate that the expressed Cas12f1 and the sgRNA from the host’s sgRNA plasmid form a Cas12f1-sgRNA complex that effectively recognizes and cleaves the galK WT and galK 504A targets, causing cell death and clear spot formation. However, in the presence of two nucleotide (nt) mismatches (galK 504AT) or the absence of the galK target (ΔgalK), the Cas12f1-sgRNA complex fails to cleave the genome, preventing cell death and allowing lysogeny to form, resulting in turbid spots (Fig. 2C).
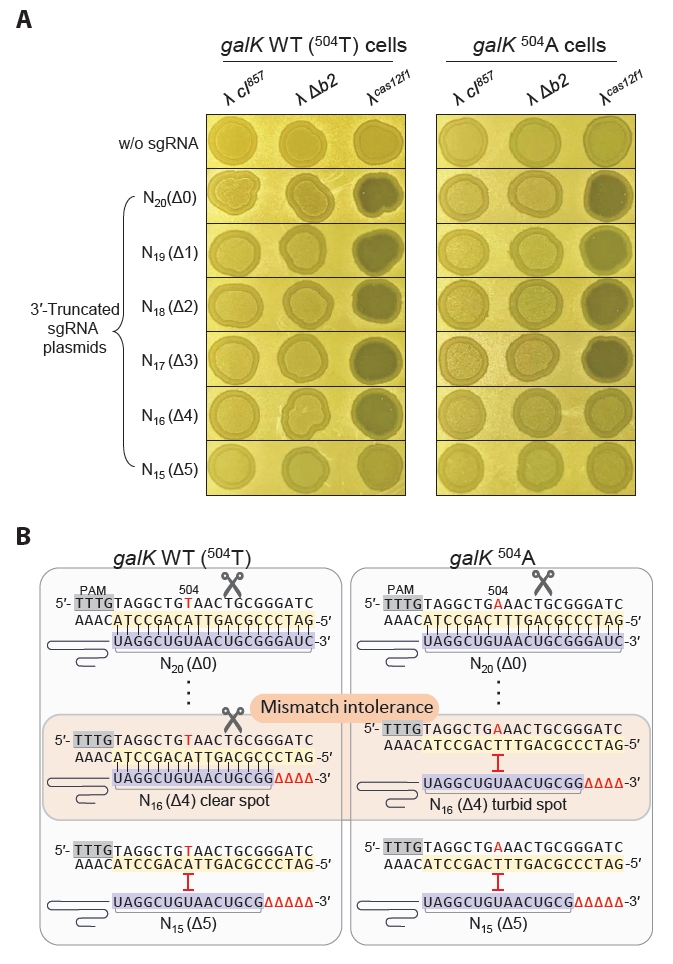
Next, to distinguish between galK WT and galK 504A, which differ by a single-nucleotide, we tested the effectiveness of a 3′-end truncated sgRNA approach (Lee et al., 2023) to overcome the mismatch tolerance of the Cas12f1 system. We performed the sameλ cas12f1 phage spotting assay using sgRNAs with TRS lengths ranging from 20 nt (Δ0) to 15 nt (Δ5). When galK WT cells harboring Δ0 to Δ4 nt sgRNA plasmids were infected with the λcas12f1 phage, clear spots were formed (Fig. 3A). However, in galK 504A cells, clear spots were only formed with Δ0 to Δ3 nt sgRNA plasmids, while Δ4 nt sgRNA plasmid-bearing cells formed turbid spots.
When the phage spotting assay was performed at 37°C, clear spots were formed on all plates, regardless of the strain or phage type (Fig. S3). These results demonstrate that when host cells are infected with a Cas12f1-bearing bacteriophage, truncation of the sgRNA enables the differentiation of single-nucleotide variations in target genes (Fig. 3B) and allows for control over host cell lysogen formation and cell death.
Genomic DNA sequence-specific lysis of E. coli via λ phage-mediated delivery of Cas12f1 and truncated sgRNAs
We engineered a synthetic phage λcas12f1-sgRNA by integrating the genes for cas12f1 and galK-targeting sgRNA into the b2 region of the bacteriophage λ genome. This synthetic phage was tested for its ability to distinguish single-nucleotide variations in the target sequence of the host genome, and its effects on lysogen formation and cell lysis were elucidated. We constructed λcas12f1galK-N20 and λcas12f1galK-N16 with TRS lengths of 20 nt (Δ0) and 16 nt (Δ4), respectively (Fig. 4A). The insertion of cas12f1 and sgRNA genes into the λ cI857 genome was confirmed using PCR, and the base sequence of the sgRNA gene was confirmed through Sanger sequencing (Fig. S4).
For the spotting assay, MG1655 WT cells mixed in soft agar plates were spotted with the engineered phages—λcas12f1galK-N20, λcas12f1galK-N16, and λ cIantisense—and incubated at 30°C for 16 h, forming a clear spot (Fig. 4B). In the soft agar of galK 504A cells, only the λcas12f1galK-N20 phage and the λ cIantisense lytic phage formed clear spots, whereas the λcas12f1galK-N16 phage formed a turbid spot. This result shows that the phage-loaded Cas12f1-truncated sgRNA (galK-N16) complex recognized and cleaved the galK WT target, preventing lysogen formation. However, λcas12f1galK-N16 did not recognize and cleave the galK 504A target.
When cultured at 30°C, the λ cIantisense lytic phage showed clear spots in galK 504AT and ΔgalK host cells. However, λcas12f1galK-N20 and λcas12f1galK-N16 formed turbid spots (Fig. 4B). This result indicates that the Cas12f1-sgRNA complex did not affect lysogen formation due to the absence of target sequence variation. All phages formed clear spots in all hosts when cultured at 37°C post-spotting (Fig. S5A).
Next, galK WT, galK 504A, 504AT, and ΔgalK host cells were infected with λ cI857, λcas12f1galK-N20, λcas12f1galK-N16, and λ cIantisense phages to monitor growth in liquid culture. In galK WT cells infected with λcas12f1galK-N20 and λcas12f1galK-N16 phages at 30°C, cell growth resumed after lysis, reaching an OD600 value of approximately 1.0 (Fig. 4C). However, in galK 504A cells, only the λcas12f1galK-N20 phage showed low growth, while cells infected with the λcas12f1galK-N16 phage reached an OD600 value of 3.8, similar to those infected with the λ cI857 phage.
Both streaking of cell cultures grown after lysis due to λcas12f1galK-N20 and λcas12f1galK-N16 phage infections in galK WT at 30°C and PCR verifications revealed that no lysogens were formed. However, five colonies derived from λcas12f1galK-N16 infections in galK 504A cells were confirmed to be lysogens (Fig. S6), consistent with the spotting assay results indicating lysogen formation by λcas12f1galK-N16 in galK 504A cells. Infections with λcas12f1galK-N20 in galK 504A cells did not result in lysogen formation, as confirmed using colony PCR. In liquid cultures of galK 504AT and ΔgalK cells infected with λcas12f1galK-N20 and λcas12f1galK-N16 phages, growth curves almost identical to those infected with the λ cI857 phage were observed (Fig. S7). This indicates that the phage-delivered Cas12f1-sgRNA complex did not recognize any target in 504AT and ΔgalK cells.
When monitoring the growth of galK WT cells and galK 504A strains at 37°C, λcas12f1galK-N20 and λcas12f1galK-N16 phages displayed growth patterns nearly identical to those of the λ cI857 phage without Cas12f1-sgRNA (Fig. S5B). This similarity is attributed to the inactivation of the heat-labile CI857, causing the phages to exclusively undergo the lytic cycle and rendering the presence of Cas12f1-sgRNA irrelevant. These results demonstrate that synthetic phages carrying both cas12f1 and sgRNA genes can effectively recognize and cleave the host cell’s target galK gene, suppressing lysogen formation. Moreover, phages carrying truncated sgRNA genes can discern single-nucleotide variations in the target DNA (Fig. 4D).
Precise control of E. coli carrying Shiga toxin genes
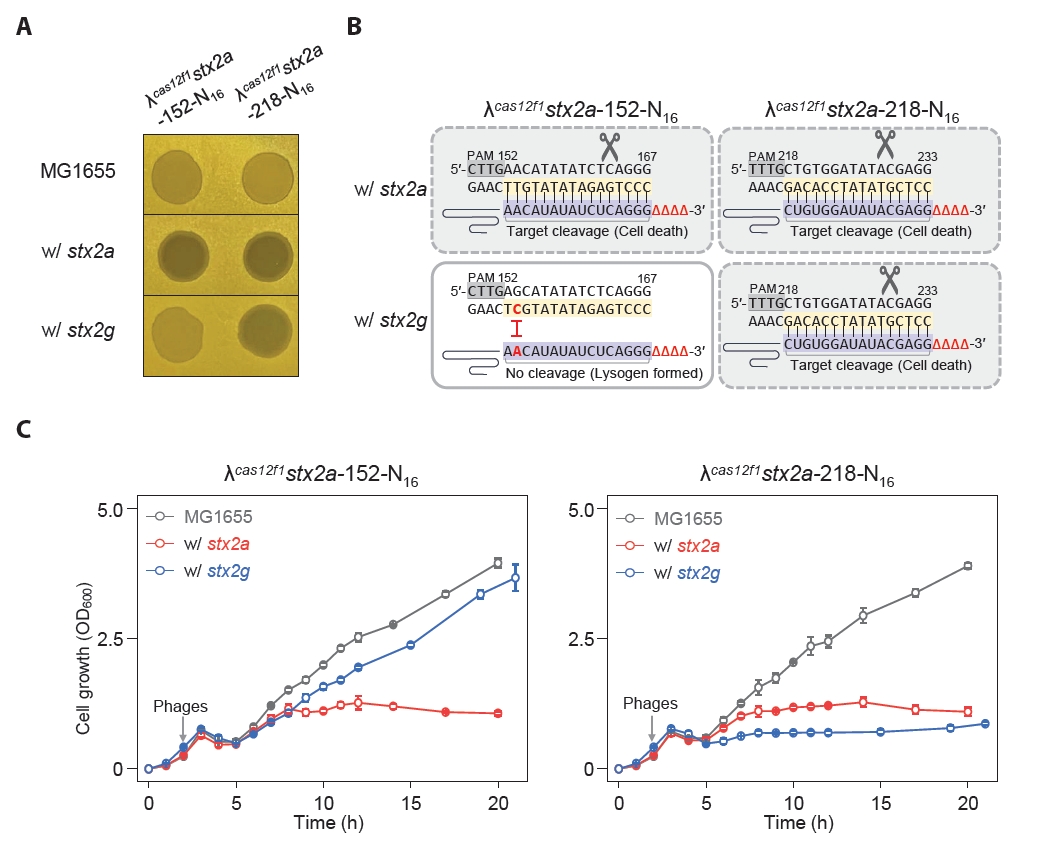
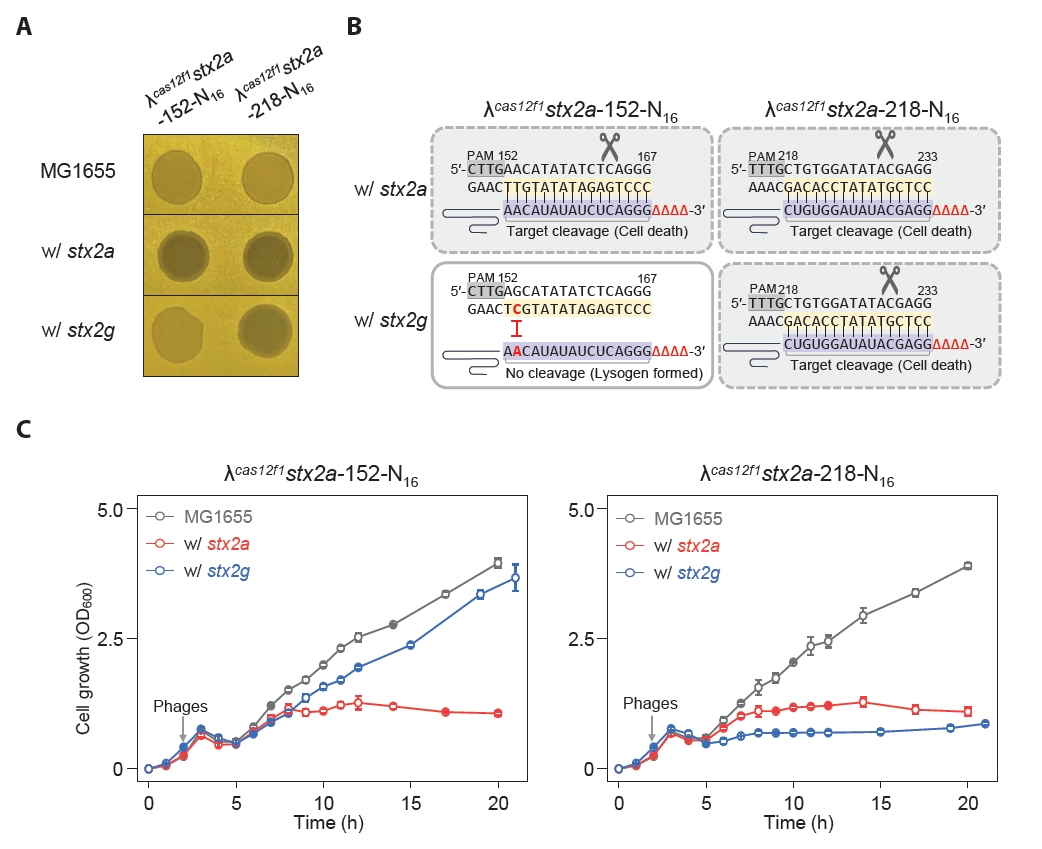
Some harmful microbes, such as Shigella and E. coli, express Shiga toxin, which can be categorized into type 1 and type 2 variants, and the subtype stx2a is known to be the most virulent (Fuller et al., 2011). We engineered MG1655 E. coli strains harboring the stx2a and stx2g genes at the srlAEBD operon location (Fig. S8). We created phages λcas12f1stx2a-152-N16 and λcas12f1stx2a-218-N16, targeting nucleotides 152–167 (16 nt) and 218–233 (16 nt) of the stx2a gene, respectively (Fig. S9). Next, we investigated the efficacy of these phages carrying truncated sgRNAs and the Cas12f1 system targeting the stx2a gene in inducing lysis and preventing lysogeny in MG1655 E. coli strains with different stx2 gene subtypes.
The spotting assay results showed that the λcas12f1stx2a-152-N16 phage formed clear spots exclusively in strains carrying the stx2a gene, while the λcas12f1stx2a-218-N16 phage formed clear spots in both stx2a and stx2g gene-carrying strains (Fig. 5A). This indicates that the λcas12f1stx2a-152-N16 phage did not recognize targets in the stx2g gene due to a single-nucleotide mismatch, thus allowing lysogen formation, as evidenced by turbid spots (Fig. 5B).
In flask culture, consistent with the spotting assay results, the λcas12f1stx2a-152-N16 phage inhibited the growth of strains carrying the stx2a gene, while strains with the stx2g gene exhibited growth curves similar to those of strains infected with the λ cI857 phage (Fig. 5C and Fig. S10). However, the λcas12f1stx2a-218-N16 phage inhibited the growth of both stx2a and stx2g gene-carrying strains due to perfect base paring between the target DNA and truncated sgRNA.
Additionally, when phages were not introduced or when infected with the λ cI857 phage, the same growth patterns were observed in all four types of strains (Fig. S10). These results demonstrate that synthetic λ phages carrying cas12f1 and truncated sgRNA can specifically control subtypes of the stx2 pathogenic genes in the host’s genome by distinguishing single-nucleotide variations.
Discussion
Prophages integrate into the host chromosome, facilitating horizontal gene transfer of antibiotic-resistance genes, pathogenic determinants, and other genetic elements (Wendling et al., 2021). When the temperate phage λ infects E. coli, cell growth is observed after lysis of the host cells, a phenomenon attributed to the formation of lysogens (Maynard et al., 2010, 2012; Sinha et al., 2017). Consistent with previous studies, infection of E. coli MG1655 cells cultured at 30°C with λ cI857 and λ Δb2 resulted in the formation of turbid spots on soft agar (Fig. 1A) and an increase in OD600 after cell lysis in liquid culture (Fig. 1B). Lambdoid prophages provide superinfection immunity to their hosts, preventing further phage infections (Fogg et al., 2010). PCR experiments confirmed that cells exhibiting post-infection growth without lysis were indeed lysogenic (Fig. S1). The formation of host cell lysogens by prophages poses a challenge to the use of phages in bacterial control strategies (Bondy-Denomy et al., 2016).
In this study, we first engineered a bacteriophage λ lacking the b2 region of its genome and equipped with the Cas12f1 nuclease. Subsequently, we characterized the λcas12f1 phage by infecting cells carrying sgRNA plasmids (Fig. 2A & 2B). The λcas12f1 phage effectively infected the host and formed clear spots only in the presence of the sgRNA. This observation highlighted the capability of the Cas12f1-sgRNA complex to inhibit lysogen formation in the host (Fig. 2C). A similar inhibitory effect on lysogeny has been reported previously with a C. difficile-specific temperate phage equipped with the CRISPR-Cas3 system (Selle et al., 2020).
Cells lacking the galK gene (ΔgalK) and those with the galK 504AT variant, which had two mismatches between the genomic galK target and sgRNA, did not facilitate targeting by the Cas12f1 nuclease-sgRNA complex, resulting in the formation of turbid spots indicative of lysogen formation. Conversely, cells with the galK 504A variant, which had only a single mismatch, were recognized as targets like galK WT (504T) cells, resulting in clear spot formation (Fig. 2B). This finding highlights a notable drawback of the CRISPR-Cas system, specifically its mismatch tolerance, where targeting occurs despite the presence of a few mismatches between the target DNA and sgRNA (Huang et al., 2022). A similar issue with Cas9 has been solved by using truncated sgRNA, which accurately identifies single-nucleotide mismatches, a phenomenon referred to as mismatch intolerance (Lee et al., 2021). Furthermore, precise genome editing has been demonstrated in the Cas12f1 system using truncated sgRNA, enabling the effective discrimination of single-nucleotide variations (Lee et al., 2023).
To evaluate and address the issue of mismatch tolerance (Fig. 2B), we performed a spotting assay using the λcas12f1 phage to infect galK WT (504T) and galK 504A cells, each carrying plasmids with truncated sgRNAs of various lengths (Lee et al., 2023). The results demonstrated that the λcas12f1 phage could effectively distinguish single-nucleotide variations in the host genome. It caused lysis in galK WT cells but allowed lysogen formation in galK 504A, where the Cas12f1-N16 sgRNA failed to recognize the target sequence (Fig. 3A). This finding underscores the capability of phage-delivered Cas12f1 to discern single-nucleotide variations within sgRNA target sequences in the host genome.
However, since the sgRNA for Cas12f1 was introduced into the host via multiple-copy plasmids, we further investigated whether single-copy sgRNA integrated into the phage genome could still effectively distinguish single-nucleotide variations. The results demonstrated that the λcas12f1galK-N16 phage, which carried both the sgRNA and Cas12f1, effectively killed galK WT cells and suppressed lysogen formation but did not kill galK 504A cells (Fig. 4A & 4B), allowing lysogen formation (Fig. S6). Thus, the λcas12f1galK-N16 phage carrying truncated sgRNA and Cas12f1 was confirmed to be capable of distinguishing single-nucleotide variations in the genome (Fig. 4B). Additionally, in liquid culture, while the λcas12f1galK-N16 phage-infected galK WT host cells initially exhibited slight growth after lysis, their growth was notably suppressed compared to the λ cIantisense lytic phage-infected host bacteria at 18 h (Fig. 4C).
After infection with the λcas12f1galK-N16 phage, galK 504A cells exhibited growth but were confirmed to be lysogens using PCR, indicating that the phage-delivered Cas12f1-truncated sgRNA complex did not recognize galK 504A as a target (Fig. S6). Conversely, as shown in Fig. 1B, cells infected with the lytic λ cIantisense phage also regrew but did not become lysogens (Fig. S1B), likely due to the emergence of phage-resistant mutants. Figure 4C illustrates that cell growth at 18 h post-infection was more suppressed by λcas12f1galK-N20 or λcas12f1galK-N16 phages than by the λ cIantisense phage, suggesting that the phage-delivered Cas12f1-sgRNA complex effectively eliminated the emerged phage-resistant mutants. Additionally, the observation of residual cell growth post-lysis by the Cas12f1-loaded λ phage, despite the absence of lysogeny, could be attributed to various factors, including the elimination, modification, or masking of phage receptors (Egido et al., 2022), emergence of persisters (Mamontov et al., 2022) and mutations in the PAM or protospacer (Schelling et al., 2023). Indeed, previous studies using the M13 phagemid to deliver the Cas9 system have reported observing escape cells due to mutations in the spacer, tracrRNA, or target site (Lam et al., 2021).
When the λcas12f1stx2a-152-N16 phage infected E. coli strains carrying different subtypes of the stx2 gene, specifically stx2a and stx2g, it could distinguish single-nucleotide variations between the two stx2 gene subtypes, effectively lysing cells with the stx2a gene (Fig. 5C). This distinction is crucial given that stx2 is known to vary in virulence depending on the subtype, with stx2a being the most virulent (Fuller et al., 2011). Such selective control over gene subtypes enhances precision in targeting specific harmful microbes within a microbial population.
Conclusion
The CRISPR-Cas system, which originally evolved as part of the microbial immune system against foreign nucleic acids from phages, is now finding applications in diverse fields, including diagnostics (Chakraborty et al., 2022) and therapeutics (Luthra et al., 2021). However, the phenomenon of mismatch tolerance has posed challenges in its applications in genome editing, diagnostics, and therapeutics (Mengstie et al., 2024). In this study, we introduced a CRISPR-Cas-loaded phage designed to detect target sequence variations with single-nucleotide precision, demonstrated within the E. coli-λ model. While comprehensive evaluation in complex environments, such as microbial communities and animal models, remains necessary, our investigation into synthetic phages represents a significant step forward in the development of precise CRISPR-Cas-based antimicrobial agents.
Acknowledgments
This work was supported by a grant from the National Research Foundation of Korea (NRF), funded by the Korean government (MSIT) (RS-2024-00342735), a grant from Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (RS-2024-00411263), and a grant from the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (HI23C0041).
Author Contributions
H.J.L.: Conceptualization, Methodology, Validation, Analysis, and Writing. S.H.J.: Methodology, Validation, Analysis, and Writing. S.J.L.: Conceptualization, Analysis, and Writing. All the authors read and approved the final version of manuscript.
Conflict of Interest
The authors have no conflict of interest to report.
Supplementary Information
The online version contains supplementary material available at https://doi.org/10.71150/jm.2501012.
Fig. S6.
Confirmation of lysogen formation in galK WT and galK504A cells infected with λcas12f1 galK-N20 or λcas12f1galK-N16 phages at 30°C and 37°C.
jm-2501012-Supplementary-Fig-S6.pdf
Fig. S10.
Growth curves of λ cI857 phage-infected E. coli cells carrying the stx2a or stx2g gene at 30°C. The gray arrows indicate the time points of phage infection.
jm-2501012-Supplementary-Fig-S10.pdf
Fig. 1.Growth of E. coli MG1655 cells infected with bacteriophages λ cI857, λ Δb2, and λ cIantisense in solid and liquid media at 30°C and 37°C. (A) Spotting assay of bacteriophages on soft agar with MG1655 cells. Turbid and clear spots represent lysogen formation and lytic cell death, respectively. (B) Growth of phage-infected cells in flask cultures at different temperatures. Gray arrows indicate the time of phage infection, and red arrows indicate the time points of sample collection for PCR. The growth measurement results represent the average values obtained from three independent cultures.
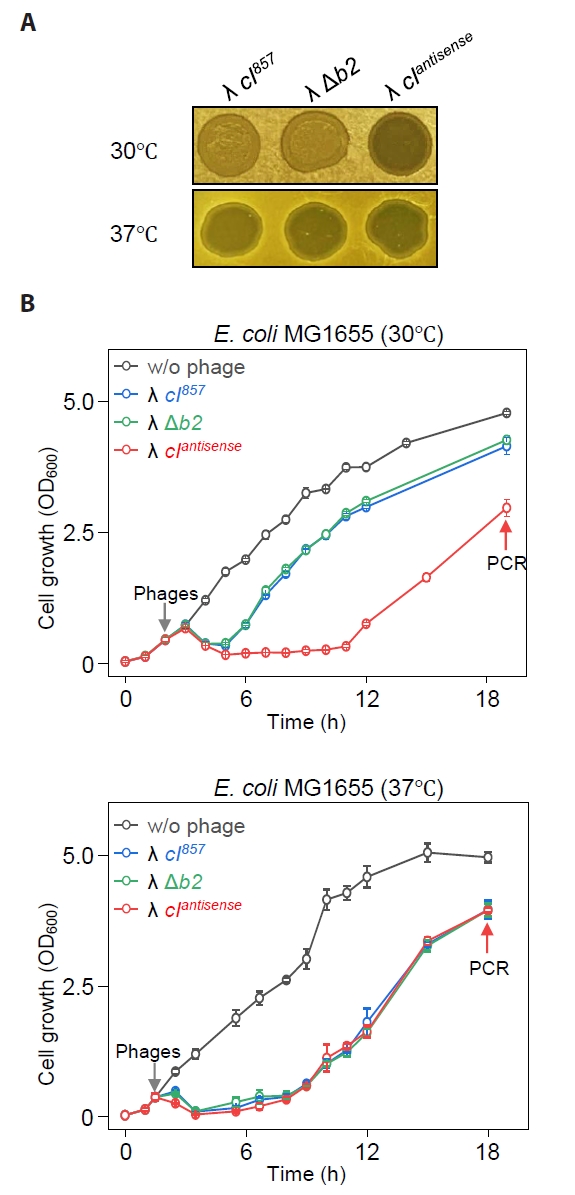
Fig. 2.Genomic target recognition and complete cell lysis by phage λ carrying Cas12f1 nuclease. (A) Construction of λcas12f1. The cas12f1 gene was inserted into the b2 region of the λ prophage genome via homologous recombination. (B) Spotting assay of λcas12f1. Various E. coli MG1655 cells with different galK genotypes (galK WT, galK 504A, galK 504AT, and ΔgalK) were tested. The cells were either transformed with or without a galK-targeting sgRNA plasmid. The phages λ cI857, λ Δb2, and λcas12f1 were spotted on soft agar containing a culture of transformed cells and incubated at 30°C for 16 h. (C) Proposed mechanism of cell death. The Cas12f1 nuclease is delivered by the external phage λ carrying the cas12f1 gene, while the sgRNA is supplied by a plasmid within the host cell. The Cas12f1-sgRNA complex cleaves host genomic DNA when the target sequence is present, causing cell death. If the target sequence is mismatched (indicated by a red asterisk) or absent, cleavage does not occur, allowing the host cell to survive and potentially enter lysogeny. This ensures that only target cells are lysed, while non-target cells remain viable.
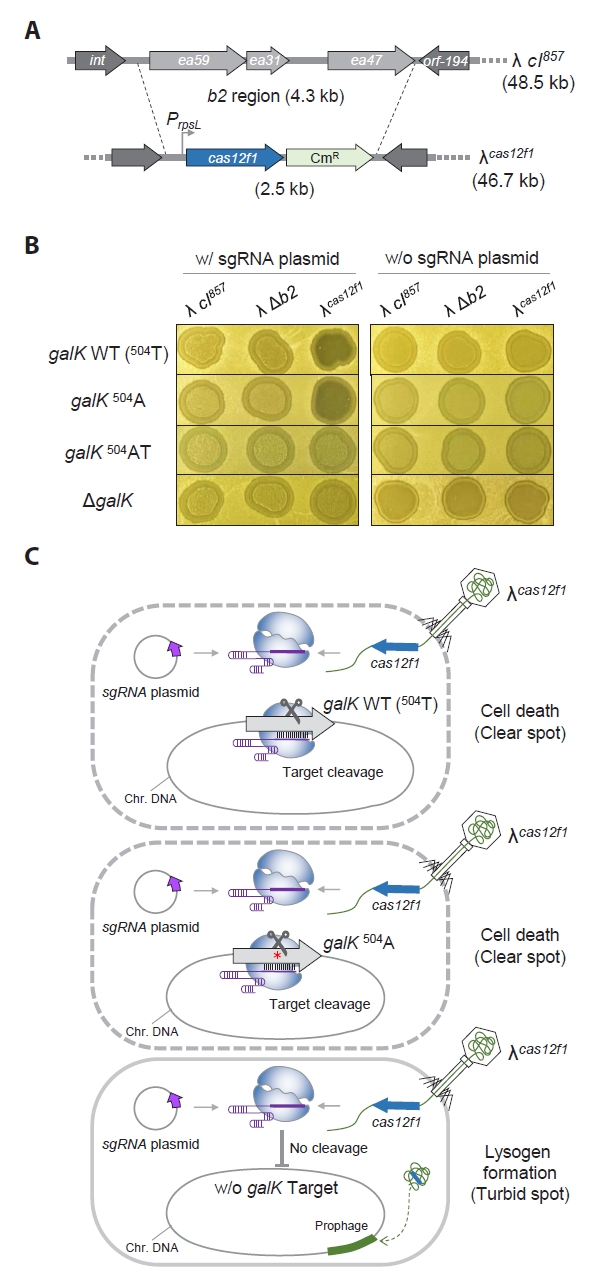
Fig. 3.
Recognition of single-nucleotide variations in the genome leveraging the mismatch intolerance of the 3ʹ-end truncated sgRNAs and phage λ-supplied Cas12f1 nuclease.
(A) Spotting assay of λcas12f1. MG1655 galK WT and 504A cells carrying different lengths of truncated sgRNAs were tested. The phages λ cI857, λ Δb2, and λcas12f1 were spotted onto soft agar containing a culture of transformed galK WT and 504A cells and incubated at 30°C for 16 h. The terms Δ0–Δ5 indicate the number of 3′-end truncations in the sgRNA, and N20–N15 denotes the length of the sgRNA target recognition sequence (TRS). (B) Mismatch intolerance. The complex formed by the 3′-end truncated sgRNAs and the Cas12f1 nuclease can discriminate single-nucleotide variations in the genomic target. 504 refers to the nucleotide position within the galK structural gene. Red nucleotides T or A indicate variations at the 504th nucleotide position. Black vertical lines represent the base pairing between the non-PAM strand of the target DNA and the sgRNA. The scissor icon indicates that the Cas12f1-sgRNA complex recognizes and cleaves the target DNA. The Δ symbol denotes truncated nucleotides in the sgRNA. The red Ⅰ symbol indicates that the target DNA sequence is not recognized by the sgRNA as a cleavage target.
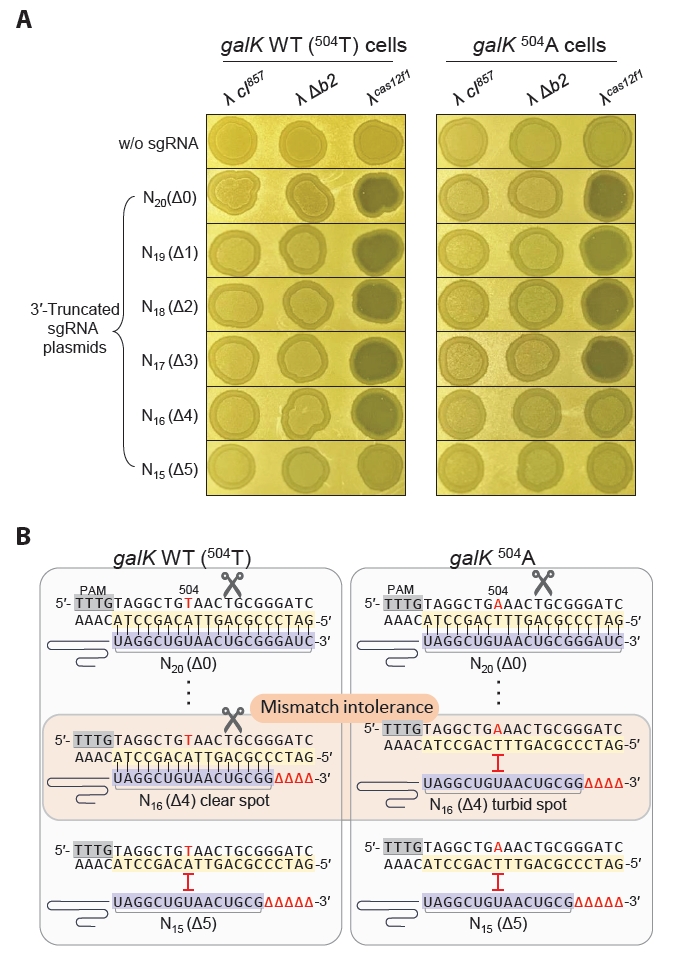
Fig. 4.Sequence-specific galK target recognition and bacterial cell control using cas12f1-sgRNA-loaded phages. (A) Construction of synthetic λ phages carrying both the cas12f1 gene and the galK-targeting sgRNA gene. Phages with cas12f1 and galK targeting sgRNA genes inserted into the λ cI857 genome were constructed. λcas12f1galK-N20 and λcas12f1galK-N16 have sgRNA TRS lengths of 20 nt and 16 nt, respectively. (B) Spotting assay of synthetic phages, including cas12f1-sgRNA-loaded λ phages, on soft agar containing MG1655, MG1655-galK 504A, MG1655-galK 504AT, or MG1655-ΔgalK cells at 30°C for 16 h. (C) Genomic galK sequence-specific bacterial control by cas12f1-sgRNA-loaded phages. The growth of the MG1655 and MG1655-galK 504A strains at 30°C was measured at OD600 after infection with λcas12f1galK-N20 and λcas12f1galK-N16 phages. The gray arrow indicates the time point of phage infection. Each OD600 measurement represents the average value obtained from three independent cultures. (D) Recognition of single-nucleotide variation and inhibition of lysogeny by λcas12f1galK-N16 phage. The λcas12f1galK-N16 phage carries a truncated sgRNA targeting the galK WT gene. The Cas12f1-truncated sgRNA complex delivered by the phage cleaves the galK WT target through perfect base pairing, thereby preventing lysogeny. However, in galK 504A cells, the genomic target is not recognized due to the presence of a single mismatch, allowing the formation of lysogenic cells.
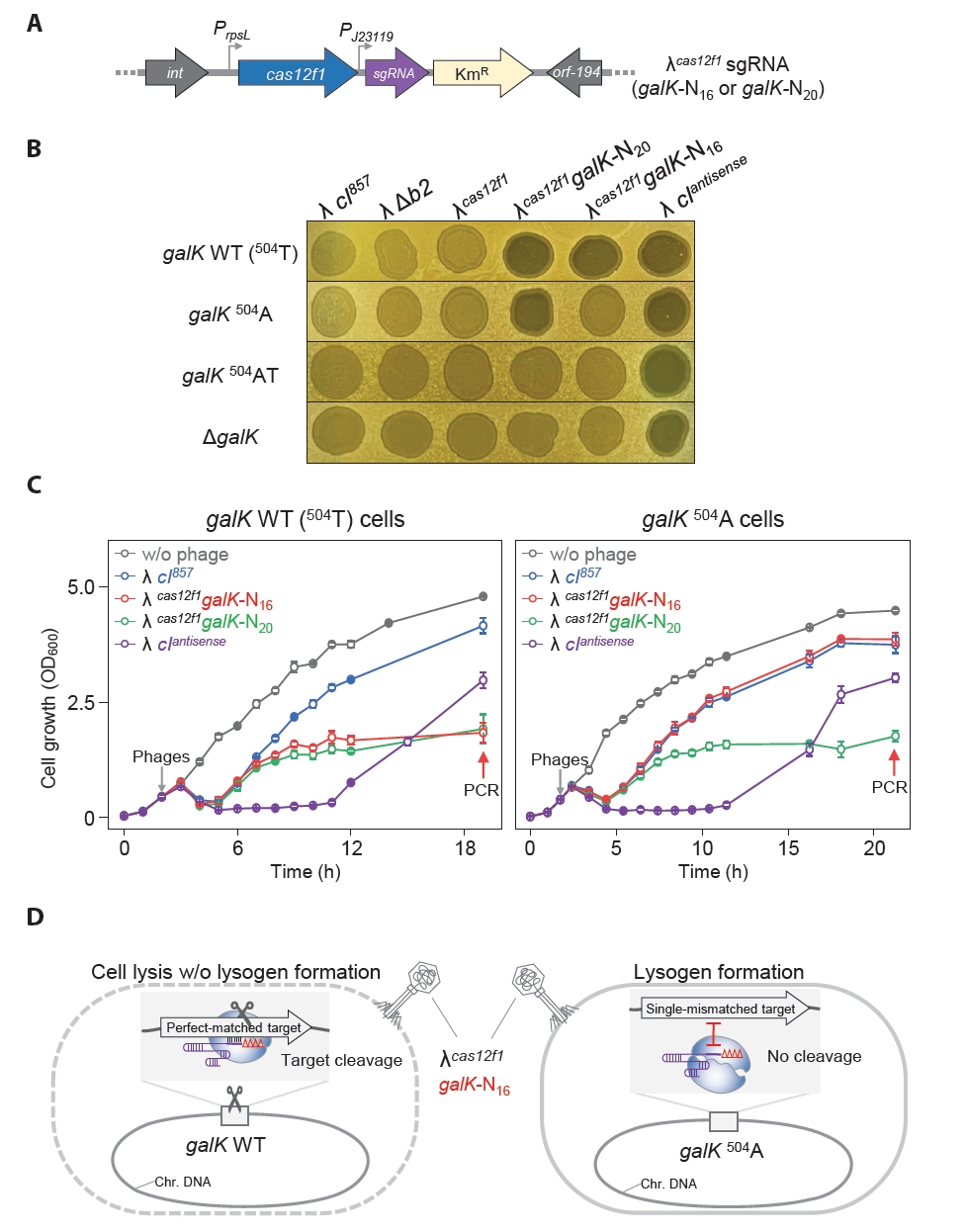
Fig. 5.Controlled lysis of E. coli carrying different Shiga toxin gene subtypes using cas12f1-sgRNA-loaded phages. (A) Spotting assay of synthetic λcas12f1stx2a-152-N16 and λcas12f1stx2a-218-N16 phages on soft agar containing various E. coli cells carrying stx2 subtypes. stx2a and stx2g genes were inserted into the srl operon of the MG1655 genome. (B) Target nucleotide sequences of stx2 gene subtypes recognized by truncated sgRNAs (N16) of the CRISPR-Cas12f1 system. Red nucleotides represent single-nucleotide variations specific to the stx2 subtypes that prevent perfect base-pairing with the sgRNA sequence. (C) Genomic stx2 subtype sequence-specific bacterial control using cas12f1-sgRNA-loaded phages. The growth of MG1655 strains carrying stx2a and stx2g genes at 30°C was monitored after infection with synthetic λcas12f1stx2a-152-N16 and λcas12f1stx2a-218-N16 phages. The gray arrow indicates the time point of phage infection. Each OD600 measurement represents the average value obtained from three independent cultures.
References
- Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, et al. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2: 2006.0008.ArticlePubMedPMC
- Bondy-Denomy J, Qian J, Westra ER, Buckling A, Guttman DS, et al. 2016. Prophages mediate defense against phage infection through diverse mechanisms. ISME J. 10: 2854–2866. ArticlePubMedPMCPDF
- Chakraborty J, Chaudhary AA, Khan SU, Rudayni HA, Rahaman SM, et al. 2022. CRISPR/Cas-based biosensor as a new age detection method for pathogenic bacteria. ACS Omega. 7(44): 39562–39573. ArticlePubMedPMCPDF
- Cobb LH, Park J, Swanson EA, Beard MC, McCabe EM, et al. 2019. CRISPR-Cas9 modified bacteriophage for treatment of Staphylococcus aureus induced osteomyelitis and soft tissue infection. PLoS One. 14(11): e0220421. ArticlePubMedPMC
- Duong MM, Carmody CM, Ma Q, Peters JE, Nugen SR. 2020. Optimization of T4 phage engineering via CRISPR/Cas9. Sci. Rep. 10: 18229.ArticlePubMedPMCPDF
- Egido JE, Costa AR, Aparicio-Maldonado C, Haas PJ, Brouns SJJ. 2022. Mechanisms and clinical importance of bacteriophage resistance. FEMS Microbiol Rev. 46(1): fuab048.ArticlePubMedPDF
- Fogg PC, Allison HE, Saunders JR, McCarthy AJ. 2010. Bacteriophage lambda: a paradigm revisited. J Virol. 84(13): 6876–6879. ArticlePubMedPMCPDF
- Fuller CA, Pellino CA, Flagler MJ, Strasser JE, Weiss AA. 2011. Shiga toxin subtypes display dramatic differences in potency. Infect Immun. 79(3): 1329–1337. ArticlePubMedPMCPDF
- Gencay YE, Jasinskyte D, Robert C, Semsey S, Martinez V, et al. 2024. Engineered phage with antibacterial CRISPR-Cas selectively reduce E. coli burden in mice. Nat Biotechnol. 42(2): 265–274. ArticlePubMedPDF
- George HJ, Watson RJ, Harbrecht DF, Delorbe WJ. 1987. A bacteriophage λ cI857 cassette controls λ PL expression vectors at physiologic temperatures. Nat Biotechnol. 5: 600–603. ArticlePDF
- Guan J, Oromi-Bosch A, Mendoza SD, Karambelkar S, Berry JD, et al. 2022. Bacteriophage genome engineering with CRISPR-Cas13a. Nat Microbiol. 7(12): 1956–1966. ArticlePubMedPMCPDF
- Hamilton TA, Pellegrino GM, Therrien JA, Ham DT, Bartlett PC, et al. 2019. Efficient inter-species conjugative transfer of a CRISPR nuclease for targeted bacterial killing. Nat Commun. 10(1): 4544.ArticlePubMedPMCPDF
- Huang XQ, Yang DS, Zhang JF, Xu J, Chen YE. 2022. Recent advances in improving gene-editing specificity through CRISPR-Cas9 nuclease engineering. Cells. 11(14): 2186.ArticlePubMedPMC
- Jia HJ, Jia PP, Yin S, Bu LK, Yang G, et al. 2023. Engineering bacteriophages for enhanced host range and efficacy: insights from bacteriophage-bacteria interactions. Front Microbiol. 14: 1172635.ArticlePubMedPMC
- Jin M, Chen J, Zhao X, Hu G, Wang H, et al. 2022. An engineered λ phage enables enhanced and strain-specific killing of enterohemorrhagic Escherichia coli. Microbiol Spectr. 10(4): e0127122. ArticlePubMedPDF
- Kim HJ, Jeong H, Lee SJ. 2021a. Visualization and quantification of genetically adapted microbial cells during preculture. Front Microbiol. 12: 693464.ArticlePubMed
- Kim DY, Lee JM, Moon SB, Chin HJ, Park S, et al. 2021b. Efficient CRISPR editing with a hypercompact Cas12f1 and engineered guide RNAs delivered by adeno-associated virus. Nat Biotechnol. 40: 94–102. ArticlePubMedPDF
- Lam KN, Spanogiannopoulos P, Soto-Perez P, Alexander M, Nalley MJ, et al. 2021. Phage-delivered CRISPR-Cas9 for strain-specific depletion and genomic deletions in the gut microbiome. Cell Rep. 37(5): 109930.ArticlePubMedPMC
- Lee HJ, Kim HJ, Lee SJ. 2021. Mismatch intolerance of 5′-truncated sgRNAs in CRISPR/Cas9 enables efficient microbial single-base genome editing. Int J Mol Sci. 22(12): 6457.ArticlePubMedPMC
- Lee HJ, Kim HJ, Lee SJ. 2022. Control of λ lysogenic Escherichia coli cells by synthetic λ phage carrying cIantisense. ACS Synth Biol. 11(11): 3829–3835. ArticlePubMedPMCPDF
- Lee HJ, Kim HJ, Lee SJ. 2023. Miniature CRISPR-Cas12f1-mediated single-nucleotide microbial genome editing using 3'-truncated sgRNA. CRISPR J. 6(1): 52–61. ArticlePubMedPMC
- Liang J, Zhang H, Tan YL, Zhao H, Ang EL. 2022. Directed evolution of replication-competent double-stranded dna bacteriophage toward new host specificity. ACS Synth Biol. 11(2): 634–643. ArticlePubMedPDF
- Lin J, Du F, Long M, Li P. 2022. Limitations of phage therapy and corresponding optimization strategies: A review. Molecules. 27(6): 1857.ArticlePubMedPMC
- Lobocka M, Dabrowska K, Gorski A. 2021. Engineered bacteriophage therapeutics: Rationale, challenges and future. BioDrugs. 35(3): 255–280. ArticlePubMedPMCPDF
- Luthra R, Kaur S, Bhandari K. 2021. Applications of CRISPR as a potential therapeutic. Life Sci. 284: 119908.ArticlePubMed
- Mamontov V, Martynov A, Morozova N, Bukatin A, Staroverov DB, et al. 2022. Persistence of plasmids targeted by CRISPR interference in bacterial populations. Proc Natl Acad Sci USA. 119(15): e2114905119. ArticlePubMedPMC
- Maynard ND, Birch EW, Sanghvi JC, Chen L, Gutschow MV, et al. 2010. A forward-genetic screen and dynamic analysis of lambda phage host-dependencies reveals an extensive interaction network and a new anti-viral strategy. PLoS Genet. 6(7): e1001017. ArticlePubMedPMC
- Maynard ND, Macklin DN, Kirkegaard K, Covert MW. 2012. Competing pathways control host resistance to virus via tRNA modification and programmed ribosomal frameshifting. Mol Syst Biol. 8: 567.ArticlePubMedPMCPDF
- Mengstie MA, Azezew MT, Dejenie TA, Teshome AA, Admasu FT, et al. 2024. Recent advancements in reducing the off-target effect of CRISPR-Cas9 genome editing. Biologics. 18: 21–28. ArticlePubMedPMC
- Mitsunaka S, Yamazaki K, Pramono AK, Ikeuchi M, Kitao T, et al. 2022. Synthetic engineering and biological containment of bacteriophages. Proc Natl Acad Sci USA. 119(48): e2206739119. ArticlePubMedPMC
- Nethery MA, Hidalgo-Cantabrana C, Roberts A, Barrangou R. 2022. CRISPR-based engineering of phages for in situ bacterial base editing. Proc Natl Acad Sci USA. 119(46): e2206744119. ArticlePubMedPMC
- Park JY, Moon BY, Park JW, Thornton JA, Park YH, et al. 2017. Genetic engineering of a temperate phage-based delivery system for CRISPR/Cas9 antimicrobials against Staphylococcus aureus. Sci. Rep. 7: 44929.ArticlePubMedPMCPDF
- Rogovski P, Cadamuro RD, da Silva R, de Souza EB, Bonatto C, et al. 2021. Uses of bacteriophages as bacterial control tools and environmental safety indicators. Front Microbiol. 12: 793135.ArticlePubMedPMC
- Schelling MA, Nguyen GT, Sashital DG. 2023. CRISPR-Cas effector specificity and cleavage site determine phage escape outcomes. PLoS Biol. 21(4): e3002065. ArticlePubMedPMC
- Selle K, Fletcher JR, Tuson H, Schmitt DS, McMillan L, et al. 2020. In vivo targeting of Clostridioides difficile using phage-delivered CRISPR-Cas3 antimicrobials. mBio. 11(2): e00019–00020. ArticlePubMedPMCPDF
- Sinha V, Goyal A, Svenningsen SL, Semsey S, Krishna S. 2017. In silico evolution of lysis-lysogeny strategies reproduces observed lysogeny propensities in temperate bacteriophages. Front. Microbiol. 8: 1386.ArticlePubMedPMC
- Sternberg SH, Doudna JA. 2015. Expanding the Biologist's Toolkit with CRISPR-Cas9. Mol Cell. 58(4): 568–574. ArticlePubMed
- Wendling CC, Refardt D, Hall AR. 2021. Fitness benefits to bacteria of carrying prophages and prophage-encoded antibiotic-resistance genes peak in different environments. Evolution. 75(2): 515–528. ArticlePubMedPMCPDF
- Yehl K, Lemire S, Yang AC, Ando H, Mimee M, et al. 2019. Engineering phage host-range and suppressing bacterial resistance through phage tail fiber mutagenesis. Cell. 179(2): 459–469.e9. ArticlePubMedPMC
- Yosef I, Manor M, Kiro R, Qimron U. 2015. Temperate and lytic bacteriophages programmed to sensitize and kill antibiotic-resistant bacteria. Proc Natl Acad Sci USA. 112(23): 7267–7272. ArticlePubMedPMC
Citations
Citations to this article as recorded by

 , Song Hee Jeong
, Song Hee Jeong , Sang Jun Lee*
, Sang Jun Lee*






 MSK
MSK

 ePub Link
ePub Link Cite this Article
Cite this Article






